Figures & data
Figure 1. The human mitochondrial genome. The inner circle represents the L-strand and the outer circle the H-strand. An expanded version of the non-coding region (NCR) is shown at the top. The displacement loop (D-loop) region contains a third strand (7S DNA), which spans between OriH and the TAS regions. The minor and major arcs of mtDNA are indicated. Abbreviations. CSB: conserved sequence block; HSP: heavy-strand promoter; LSP: light-strand promoter; OriH: heavy-strand origin; OriL: light-strand origin; TAS: termination-associated sequence.
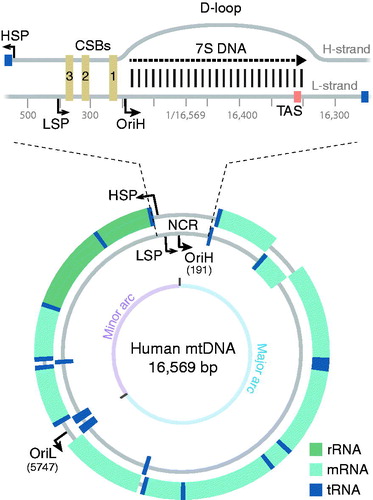
Figure 2. Schematic figure of the mtDNA replication fork. The TWINKLE DNA helicase (light blue) travels on the parental H-strand in the 5′ to 3′ direction while unwinding dsDNA. The mtSSB protein (green) binds to ssDNA and stimulates POLγ (dark blue)-dependent synthesis of the nascent H-strand. POLγ also performs L-strand DNA synthesis, using the displaced, parental H-strand as a template. POLRMT (purple) synthesizes the RNA primer (orange) needed for the initiation of L-strand synthesis at OriL. See the color version of this figure at www.tandfonline.com/ibmg
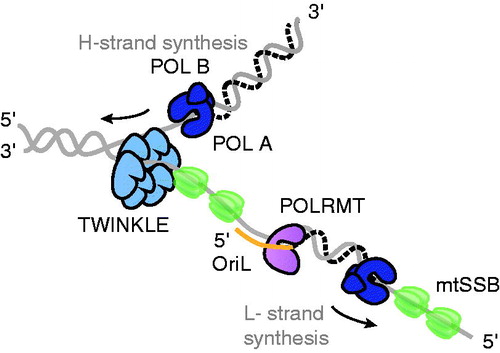
Figure 3. Strand-displacement mtDNA replication. Replication of the nascent H-strand is initiated at OriH and proceeds unidirectionally. In the process, the parental H-strand is displaced and bound by mtSSB (green), which prevents unspecific primer formation by POLRMT (purple). When the replication machinery reaches OriL, the H-strand of the origin folds into a stem-loop structure. POLRMT (purple) initiates RNA synthesis from the poly-dT stretch in the loop region, leading to the production of a short primer that is used to initiate L-strand DNA synthesis. The nascent L-strand is synthesized continuously until full-circle and two new full-length circular daughter molecules are formed. Please note that TWINKLE (light blue) is not needed for L-strand synthesis since the parental H-strand used as a template is already single-stranded. Synthesis of the daughter molecule containing the nascent H-strand is initiated and terminated at OriH, whereas synthesis of the other daughter molecule is initiated and terminated at OriL. See the color version of this figure at www.tandfonline.com/ibmg
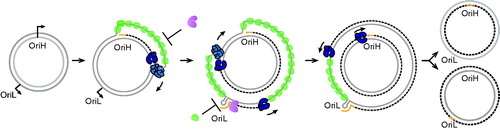
Figure 4. A schematic model for replication initiation at OriH. POLRMT initiates transcription at LSP. During transcription (yellow line) of the G-rich CSB2 region, a hybrid G-quadruplex (G4) structure is formed between RNA and the non-template H-strand. The G4 structure anchors the RNA to DNA, forming a stable R-loop. The G4 structure also causes premature transcription termination immediately downstream of CSB2 (yellow arrow). The RNA 3′ end in the nascent R-loop is not accessible to POLγ. To act as a primer, the R-loop needs to be processed by RNase H1 (scissors) This generates 3′-ends (yellow dot lines), from which POLγ can initiate DNA synthesis (black dot arrows). Both R-loop formation and DNA replication initiation are stimulated by mtSSB (green). Light brown squares; CSB 1, 2, and 3. See the color version of this figure at www.tandfonline.com/ibmg
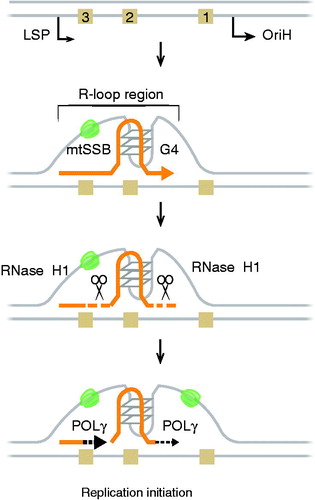
Figure 5. Primer removal at OriH and OriL. (A) At OriL, the RNA primer (∼25 nt, dashed orange line) is removed by RNase H1 leaving behind 1–2 ribonucleotides attached to the 5′-end of the nascent L-strand. Replicating POLγ displaces the 5′-end during the completion of L-strand DNA synthesis, and the last ribonucleotides are removed by a FEN1-like activity. Once a ligatable nick is produced, it is sealed by Ligase 3. (B) At OriH, the 5′-end of the nascent H-strand is processed and shifted from CSB3/CSB2 to OriH at position 191. In the process, primer RNA and a stretch of nascent DNA (∼100 nts) are removed. The 5′-end maturation process is independent of genome length mtDNA synthesis, since the 5′-end of the short 7S DNA is also processed. During primer removal, RNase H1 removes the primer (dashed orange line), leaving behind 1–2 ribonucleotides attached to the 5′-end of the nascent H-strand. In the next step, MGME1 removes these remaining ribonucleotides together with a stretch of nascent H-strand DNA (dashed red line). MGME1 works on a single-stranded DNA flap, but how the flap structure is formed is not known. See the color version of this figure at www.tandfonline.com/ibmg
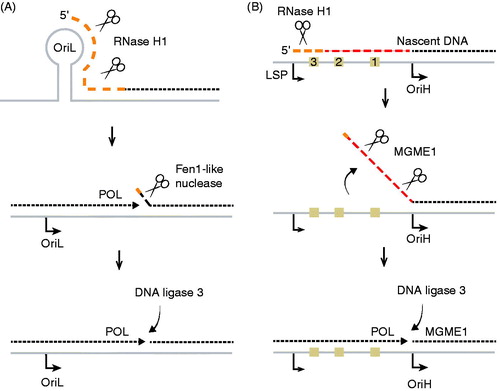
Figure 6. The role of TOP3A in the resolution of mtDNA molecules. Replication of circular mtDNA generates daughter molecules linked together by a hemicatenane structure in the OriH region. Interlinked daughter molecules are resolved by TOP3A (top panel). Lack of TOP3A activity causes the formation of catenated molecules that are unable to segregate correctly.
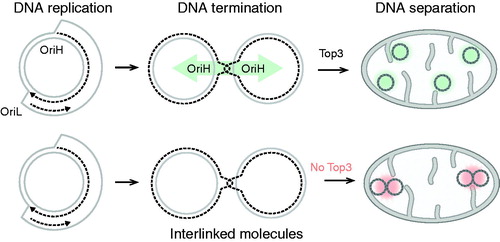
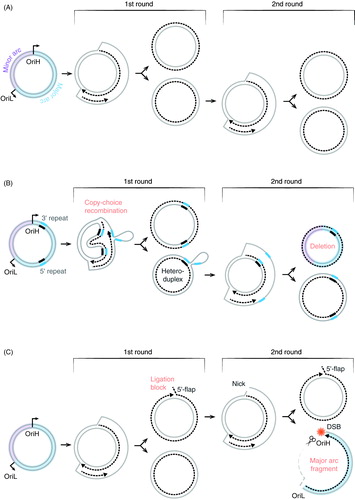
Figure 7. A model for how copy-choice recombination can lead to the formation of deleted mtDNA molecules. (A) Strand displacement of DNA replication. (B) Formation of circular deletions. During strand displacement DNA replication, repeat sequences in the parental H-strand are exposed. If POLγ dissociates from the template during replication of the first repeat, the 3′-end of the newly synthesized DNA may unpair from the template H-strand and mispair with a second repeat, located further downstream on the same strand. When POLγ resumes DNA synthesis from the mispaired 3′-end, the repeat closes to OriH is lost together with the intervening sequence. In this way, a heteroduplex molecule is formed, which when it is used as a template for the second round of mtDNA synthesis, generates one full length and one deletion-containing mtDNA molecules. Copy-choice recombination only requires the mitochondrial replication machinery and it is independent of repair mechanisms. The process may be stimulated by secondary structure elements in the H-strand or disease-causing mutations leading to reduced replication processivity. (C) Formation of linear fragments during mtDNA synthesis. Disease-causing mutations in MGME1 or loss of the POLγ 3′ to 5′ exonuclease activity can impair the formation of ligatable ends at OriH, leaving a nick in the H-strand (1st round). During the next round of mtDNA replication (2nd round), the initial phase of H-strand synthesis can proceed undisturbed, since the template L-strand is intact. However, DNA synthesis initiated at OriL will use the nicked H-strand as template and replication will therefore be prematurely terminated near OriH. As a result, a linear, deleted fragment will be formed. The ssDNA region of the fragment will be degraded, leaving a linear double-stranded product covering the entire major arc (right lower panel). Failure to ligate at OriH will therefore lead to the formation of two different replication products. One linear, double-stranded fragment and one circular mtDNA molecule with a nick at OriH. If the nicked molecule is used for a new round of mtDNA replication, the same two products may form again. Abbreviations: DSB, double-strand break.