Figures & data
Figure 1. Membrane-bound α-syn structure (2kkw.pdb) (Rao et al. Citation2010) showing its three regions, N-term (positively charged), the NAC (highly hydrophobic), and the C-term (negatively charged). The last amino acid of the N-term domain (Lys-60) and the first amino acid of the C-term domain (Lys-96), encompassing the NAC, are shown as vdW spheres. The chain colors correspond to the following secondary structures: α-helix (orange), turn (yellow), and coil (ice blue).
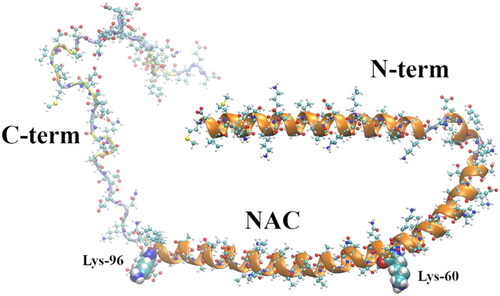
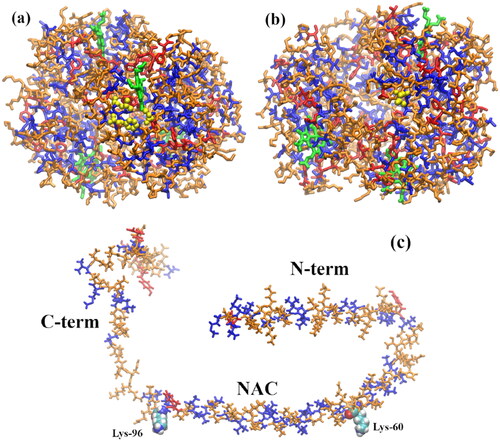
Figure 2. (a) α-syn monomer extracted from the α-syn experimental (NMR spectroscopy) protofibril reported by Tuttle et al. (Citation2016) (2n0a.pdb). (b) α-syn dimer extracted from the same experimental protofibril; cartoon representation showing a β-sheet-rich region in the NAC (amino acids 61–95) domain; some β-sheet is also visible in the N-term region. The last amino acid of the N-term domain (Lys-60) and the first amino acid of the C-term domain (Lys-96), encompassing the NAC, are shown as vdW spheres. The chain colors correspond to the following secondary structures: 310 helix (orange), β-sheet (red), turn (yellow), and coil (ice blue).
Figure 3. (a) Normal deoxygenated (T-state) hemoglobin, deoxy-HbA (2dn2.pdb) (Park et al. Citation2006), showing the four heme groups and the Glu-β6 amino acid in β1 and β2, replaced by Val-β6 in HbS; (b) sickle-cell deoxy-HbS dimer (2HBS.pdb) (Harrington et al. Citation1997) showing the Val-β6 (grey spheres) in 2β2, lodged in a hydrophobic cavity in 1β1 formed by Ala-β70, Phe-β85, and Leu-β88 (blue spheres) (Wishner et al. Citation1975; Dykes et al. Citation1979; Padlan and Love Citation1985a, Citation1985b).
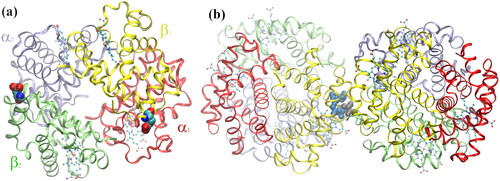
Figure 4. (a) Glu-β6 (HbA) and (b) Val-β6 (HbS) interactions with every residue from adjacent HbA and HbS tetramers, respectively. The residues forming the hydrophobic pocket in HbS-1, Ala-β70, Phe-β85, and Leu-β88, are shown in orange in (b). The most important residues in the 1β1 (blue) and Glu-β6 and Val-β6 in the 2β2 (red) polypeptides from a MD snapshot are shown below. The potential energy, and not the electrostatic and vdW energy, are plotted in (a) and (b), respectively. Reprinted from Galamba (Citation2019).
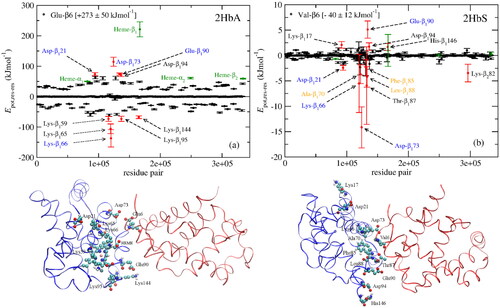
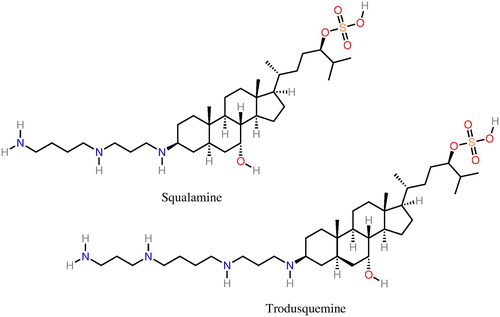
Figure 5. Molecular structure of several α-syn aggregation-inhibitors: dopamine, OleA, EGCG, Curcumin, SynuClean-D, and Fasudil.
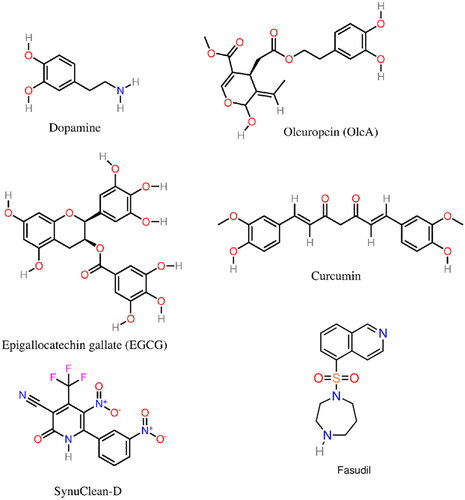
Table 1. Examples of α-syn aggregation inhibition peptides.
Figure 7. Molecular structure of several α-syn deoxy-HbS aggregation inhibitors: n-propyl urea, benzyl alcohol, clofibric acid, and gemfibrozil.
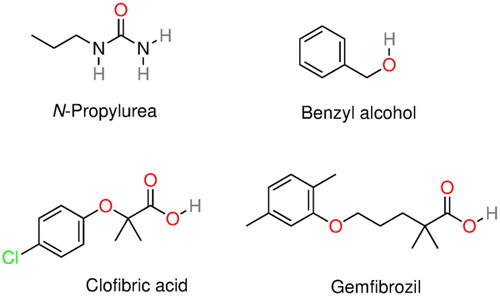
Figure 8. Molecular structure of several α-syn deoxy-HbS aggregation inhibitors: phenylalanine, tryptophan, alizarin, piperine, capsaicin, and cubebin molecular structures. Phenylalanine and tryptophan exhibit some aggregation inhibition activity and were used as building blocks in aggregation inhibition peptides (Dean and Schechter Citation1978a; Schechter et al. Citation1978). Alizarin (hydroxyl anthraquinone) is a bioactive compound from the plant Rubia cordifolia. Piperine, capsaicin, and cubebin have been pointed out (Ameh et al. Citation2012) as possible aggregation inhibitors present in Niprisan (Iyamu et al. Citation2002) (drug Nix-0699) a product of the extracts of four different plants.
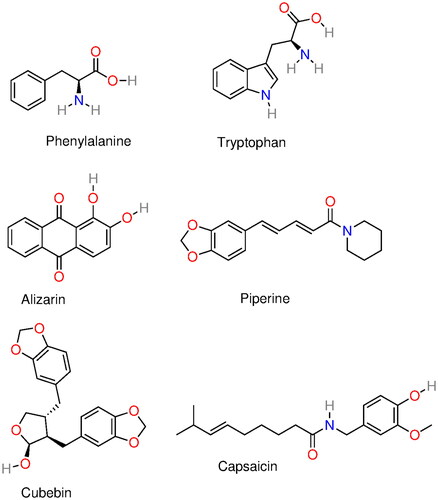
Table 2. Set of 50 compounds, among the 106, reported by Metaferia et al. (Citation2022) with antisickling activity (see Table 1 of Metaferia et al. Citation2022 for additional details).
Figure 9. (a) A water pentamer next to Ile-6 in an 11-mer isoleucine peptide (1ILE11) displaying a water in the Cβ coordination sphere with at least four nearest water neighbors (W-4W) and a water with three or less nearest water neighbors (W-3W) because of the proximity of the solute; (b) number of water molecules in the first hydration layer (rmin ≤ 6.45 Å) of the Cβ of, Ile-6 in an 11-mer isoleucine peptide (1ILE11), Ile-6 in an 11-mer serine peptide with a single (middle) isoleucine (1SER5-ILE-7SER11), and Ile-4 in NAC-term (85AGSIAAATGFV95), an 11-mer peptide comprised of the last 11 amino acids of NAC; (c) tetrahedrality of the W-4W water populations in the first hydration shell of the Cβ of Ile and Ser, respectively, in isoleucine (1ILE11) and serine (1SER11) 11-mer peptides, compared with the tetrahedrality of bulk water; (d–g) water pair interaction energy distributions, P(W···Wn) for n = 1–4, for bulk water and W-4W and W-3W water populations in the first hydration shell of the Cβ of Ile in isoleucine (1ILE11).
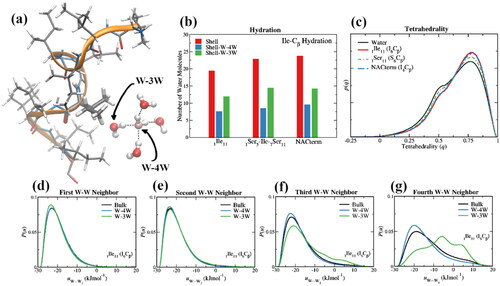
Figure 10. (a) Deoxy-HbS monomer showing the hydrophobic pocket (yellow); (b) deoxy-HbS monomer showing Val-β6 (yellow); (c) α-syn. (blue) Hydrophobic amino acids with aliphatic side chains (Ala, Ile, Leu, Met, and Val); (red) hydrophobic amino acids with aromatic side chains (Phe, Trp, Tyr); (orange) remaining amino acids; (green) Hem in deoxy-HbS.