Figures & data
Figure 1. Conventional chromosome and FISH results. (A) Representative karyogram displaying a complex karyotype with a derivative chromosome 9 with inserted material from chromosomes 11 and X (arrow), monosomy 22, trisomy 10, and material of indeterminate origin at 11q13 and at 12p11.2. (B, C) Interphase nuclei demonstrating one fusion (B) and two fusion signals (C) involving KMT2A and MLLT3 per nucleus (arrow) with dual color, dual fusion FISH. (D) Metaphase nucleus illustrating derivative chromosomes 9 (black dashed arrow) and 11 (full black arrow) with a KMT2A::MLLT3 fusion and an uninvolved chromosome 11 (white arrow) with metaphase FISH. (E) Interphase nucleus demonstrating an ETV6::RUNX1 fusion (arrows) with dual color, dual fusion FISH.
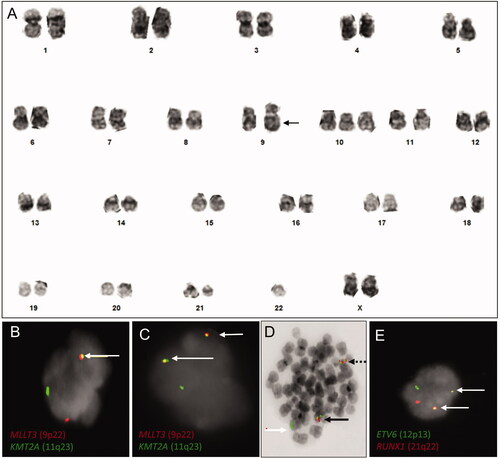
Figure 2. Schematic representation of genomic events involving KMT2A, MLLT3, CDKN2A, and ETV6::RUNX1 as identified by whole genome sequencing (WGS). (A) Representation of the ETV6::RUNX1 fusion [Citation10]. (B1) Representation of the nonproductive KMT2A rearrangement resulting from an insertion of KMT2A (from intron 1) 470 kb upstream of MLLT3 and leading to the ‘false-positive’ fluorescence in situ hybridization (FISH) fusion signal. (B2) Genomic location of breakpoints on chromosomes 9 and 11. (C1) Illustration of normal chromosomes 9p and 11q with genomic regions labeled A through F, chromosomal microarray results and Normalized Read Depth (NRD) scores. (C2) Representation of the three likely subclonal malignant populations and the corresponding FISH signal patterns. From a clone alpha (α) with an insertion of a genomic sequence from chromosome 11 (including part of KMT2A) and a deletion involving CKDN2A on the derivative chromosome 9, two subclones (beta (β) and gamma (γ)) with homozygous loss of CDKN2A likely emerged (with copy neutral loss of heterozygosity (cnLOH) from endoreduplication of the derivative chromosome 9 in the β subclone (leading to the two apparent fusion signals by FISH) and from an additional deletion involving CDKN2A on the other chromosome 9 in the γ subclone). Schematic representations were created using ProteinPaint [Citation10].
![Figure 2. Schematic representation of genomic events involving KMT2A, MLLT3, CDKN2A, and ETV6::RUNX1 as identified by whole genome sequencing (WGS). (A) Representation of the ETV6::RUNX1 fusion [Citation10]. (B1) Representation of the nonproductive KMT2A rearrangement resulting from an insertion of KMT2A (from intron 1) 470 kb upstream of MLLT3 and leading to the ‘false-positive’ fluorescence in situ hybridization (FISH) fusion signal. (B2) Genomic location of breakpoints on chromosomes 9 and 11. (C1) Illustration of normal chromosomes 9p and 11q with genomic regions labeled A through F, chromosomal microarray results and Normalized Read Depth (NRD) scores. (C2) Representation of the three likely subclonal malignant populations and the corresponding FISH signal patterns. From a clone alpha (α) with an insertion of a genomic sequence from chromosome 11 (including part of KMT2A) and a deletion involving CKDN2A on the derivative chromosome 9, two subclones (beta (β) and gamma (γ)) with homozygous loss of CDKN2A likely emerged (with copy neutral loss of heterozygosity (cnLOH) from endoreduplication of the derivative chromosome 9 in the β subclone (leading to the two apparent fusion signals by FISH) and from an additional deletion involving CDKN2A on the other chromosome 9 in the γ subclone). Schematic representations were created using ProteinPaint [Citation10].](/cms/asset/18053a05-208d-44ad-82e4-0ac2de0792b7/ilal_a_2064991_f0002_c.jpg)
