Figures & data
Figure 1. List of the first 10 SNPs found to be associated with CLL risk and the association with CLL and MBL risk across five studies. InterLymph CLL study by Berndt et al. including 4486 CLL and 13,183 controls (Black); AA CLL study by Kleinstern et al. including 173 AA CLL cases from the AA CLL cohort and 235 AA controls from the Mayo Clinic Biobank (red); MBL meta-analysis study by Crowther-Swanepoel et al. including 419 individuals with MBL and 1753 controls obtained from three sites (blue); GEC MBL study by Kleinstern et al. including 95 familial MBL individuals and 1,267 Mayo controls (blue); Mayo Clinic MBL study by Kleinstern et al. including 560 individuals with MBL from the Mayo Clinic and 2631 controls from the Mayo Clinic Biobank (blue); SNPs: single nucleotide polymorphisms; CLL: chronic lymphocytic leukemia; MBL: monoclonal B-cell lymphocytosis; InterLymph: International Lymphoma of Epidemiology Consortium; AA: African ancestry; GEC: genetic epidemiology of CLL Consortium; OR: odds ratio; CI: confidence interval.
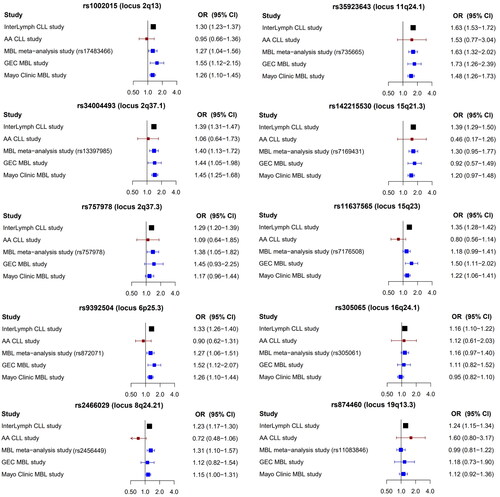
Figure 2. List of additional 31 SNPs found to be associated with CLL risk and the association with CLL and MBL risk across four studies. InterLymph CLL study by Berndt et al. including 4,486 CLL and 13,183 controls (Black); AA CLL study by Kleinstern et al. including 173 AA CLL cases from the AA CLL cohort and 235 AA controls from the Mayo Clinic Biobank (red); GEC MBL study by Kleinstern et al. including 95 familial MBL individuals and 1267 Mayo controls (blue); Mayo Clinic MBL study by Kleinstern et al. including 560 individuals with MBL from the Mayo Clinic and 2631 controls from the Mayo Clinic Biobank (blue); SNPs: single nucleotide polymorphisms; CLL: chronic lymphocytic leukemia; MBL: monoclonal B-cell lymphocytosis; InterLymph: International Lymphoma of Epidemiology Consortium; AA: African ancestry; GEC: genetic epidemiology of CLL Consortium; OR: odds ratio; CI: confidence interval.
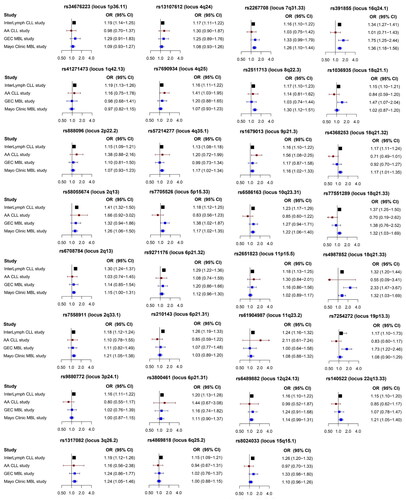
Table 1. List of 41 SNPs associated with CLL risk and their association with MBL risk across three studies.
Figure 3. (A) Distribution of the CLL-PRS by controls and CLL/MBL cohorts from different studies. Boxplots representing the CLL-PRS distribution based on 41 CLL susceptibility SNPs, across controls (light grey), AA controls (dark grey), individuals with MBL (blue), CLL cases (red), and AA CLL cases (dark red). The horizontal line and score in each box represent the median CLL-PRS for each cohort. (B) Forest plot of the association between the CLL-PRS and risk of CLL and MBL across different studies. *InterLymph study by Kleinstern et al. including 8 studies from InterLymph with a CLL-PRS based on 41 CLL-susceptibility SNPs; **InterLymph study by Berndt et al. (in Press, Leukemia) including 24 studies from InterLymph with a CLL-PRS based on 43 CLL-susceptibility SNPs; PRS: polygenic risk score; SNPs, single nucleotide polymorphisms; CLL: chronic lymphocytic leukemia; MBL: monoclonal B-cell lymphocytosis; LC: low-count; HC: high-count; AA: African ancestry; InterLymph: International Lymphoma of Epidemiology Consortium; GEC: genetic epidemiology of CLL Consortium; FH: family history; OR: odds ratio; CI: confidence interval.
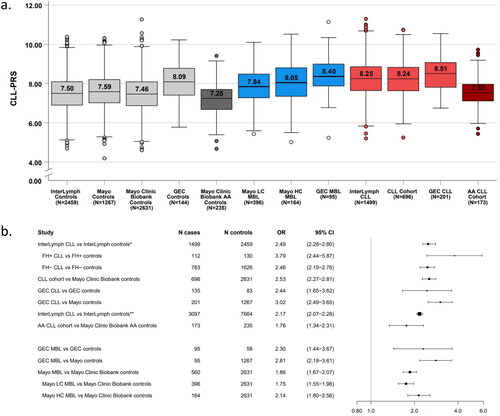
Table 2. Cohort and model description of the polygenic risk score studies.
