Figures & data
Figure 1. Clinical presentation of the patient with the CNTRL::FGFR1 fusion. (A) Schematic representation of the treatment course of the patient with the CNTRL::FGFR1 fusion. (B) Peripheral WBC counts, ANC, PLT, peripheral blood and bone marrow blast cell percentage, FISH positive signal before and during treatment with olverembatinib. (C) QRT-PCR analysis from patient bone marrow samples before and during therapy with olverembatinib. A fragment of a 635 bp unique CNTRL::FGFR1 fusion sequence was amplified. A mock RT-PCR reaction with water was included as controls. (D) Cytogenetic analyses showed the reciprocal translocation t(8;9) (p11.2;q33.2) and complex abnormalities. **p < 0.01. PR: partial remission; McyR: major cytogenetic response; WBC: white blood cell; ANC: absolute neutrophil counts; PLT: platelet counts; FISH: fluorescence in situ hybridization; QRT-PCR: Quantitative real-time polymerase chain reaction.
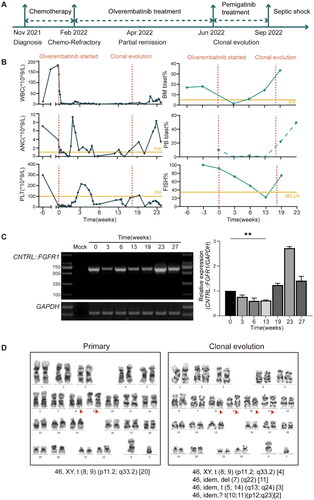
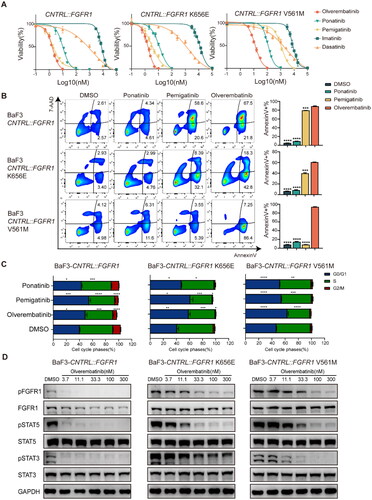
Figure 2. Olverembatinib inhibits cell function in Ba/F3 cells expressing CNTRL::FGFR1. (A) BaF3-CNTRL::FGFR1, CNTRL::FGFR1K656E, CNTRL::FGFR1V561M exhibited differential sensitivity to TKIs. (B) Olverembatinib induced significant apoptosis in CNTRL::FGFR1 expressing cells after 48 h. BaF3-CNTRL::FGFR1, CNTRL::FGFR1K656E were treated with 10 nM of olverembatinib, ponatinib and pemigatinib, while BaF3-CNTRL::FGFR1V561M was treated with 100 nM of the three drugs. (C) Effect of olverembatinib, ponatinib and pemigatinib on cell cycle distribution in Ba/F3 cells after 24 h. The concentration of the drugs is the same as apoptotic assays. (D) Olverembatinib dose-dependently inhibits phosphorylation of FGFR1, STAT5 and STAT3. Cells were treated with the olverembatinib at the different concentrations for 4 h. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Supplemental Material
Download MS Word (1.5 MB)Data availability statement
The data that support the findings of this studc from the corresponding author upon reasonable request.
