Figures & data
Table 1. Summary of sequence data of all four buffalo breeds.
Table 2. Summary of sequencing and mapping results of all four buffalo breeds.
Figure 1. (a) Venn diagram depicting common and unique InDels in all four breeds of buffalo. (b) Exome-wide probability of InDels detection in four breeds of buffalo. The content of the buffalo reference genome sequence was represented by the outermost circle. Proceeding from outer to inner of the circle showed the exome-wide spreading of InDels in Chhattisgarhi, Gojri, Murrah, and Chilika, respectively.
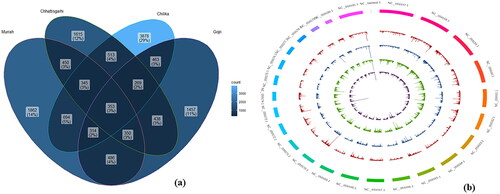
Figure 2. Population structure and relationships of Chhattisgarhi, Chilika, Gojri and Murrah buffaloes. (a) The optimal number of clusters present in the analyzed buffalo germplasms was estimated using the Evanno technique employing estimates of the number of sub-populations (K) using various statistics. (b) Buffalo breeds model-based clustering with K = 2 to 4 using the STRUCTURE programme. (c) Using principal component analysis (PCA), the comparison of PC1 and PC2 and PC3 is shown.
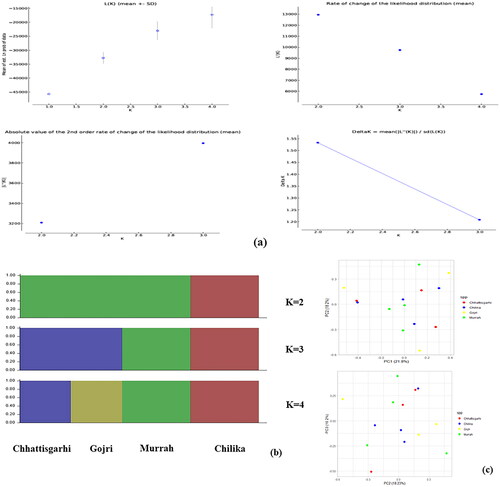
Figure 3. Summary statistics for population genetics. (a) Histogram showing frequency of different nucleotide diversity values throughout chromosome. Summary statistics for population genetics. (b) Box plots displaying distribution of nucleotide diversity, measured exome-wide across 100-kb windows in all four breeds. (c) Histogram showing frequency of different Tajima’s D values throughout exome.
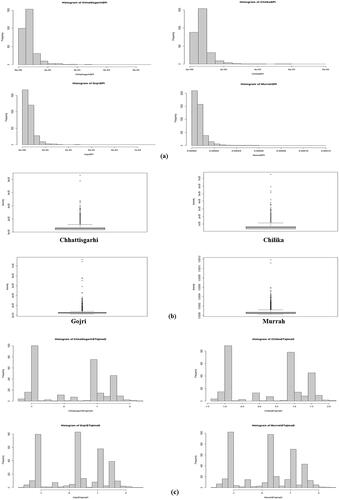
Table 3. The summary of nucleotide diversity in all four buffalo breed populations.
Figure 4. Whole exome distribution of ZFST values estimated in 100-kb sliding windows with 25-kb increments in all four breeds of buffalo.
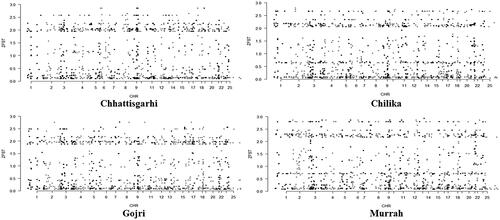
Table 4. Summary of genes identified for performance traits in all four studied breeds.
Figure 5. (a) Summary of the top 10 GO-slim biological processes into which the candidate genes were classified. (b) Network analysis of genes identified in present study in all four breeds.
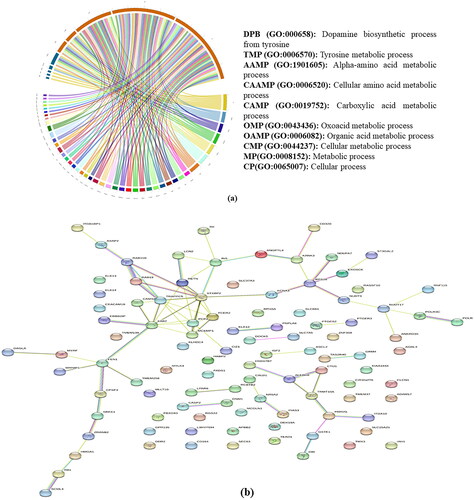
Table 5. Top 10 genes based on the number of interactions identified in the STRING database.
