Figures & data
Table 1. Detail information of PCR primers used in the study with the corresponding amplicon size and annealing temperature.
Figure 1. The relative expression of miR-361-5p in the SHF bugle and stem cells before and after activation with its functional significance in the induced activation of SHF-stem cells of cashmere goats. (A) Relative expression of miR-361-5p in cashmere goat SHF bugle at anagen and telogen stages. (B) Relative expression of miR-361-5p before and after induced activation of SHF-stem cells. (C) Overexpression efficiency analysis of miR-361-5p in SHF-stem cells of cashmere goats. (D) Overexpression of miR-361-5p led to the significant decreasing expression of three indictor genes in SHF-stem cells including Ki67, Sox9 and CD200. (E) Konckdown efficiency analysis of miR-361-5p in SHF-stem cells of cashmere goats. (F) Konckdown of miR-361-5p led to the significant increasing expression of three indictor genes in SHF-stem cells including Ki67, Sox9 and CD200. The ‘*’, ‘**’ and ‘***’ stand for indicating significant difference with p < 0.05, p < 0.01 and p < 0.001, respectively.
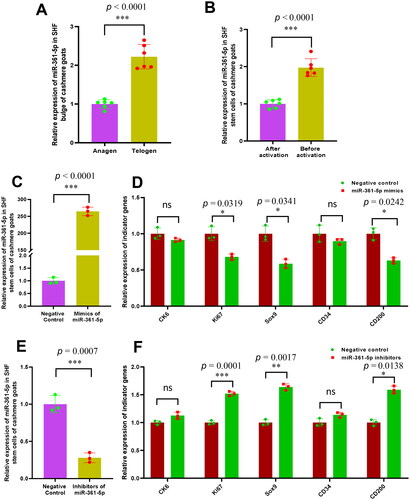
Figure 2. MiR-361-5p directly binds with FOXM1 mRNA and negatively regulates its expression in SHF-stem cells of cashmere goats. (A) Conservative analysis of miR-361-5p mature sequences among mammal species. (B) Relative expression of FOXM1 mRNA in cashmere goat SHF bugle at anagen and telogen stages. (C) Relative expression of FOXM1 mRNA before and after induced activation of SHF-stem cells. (D) A diagram of goat FOXM1 mRNA with the prediction of binding sites of miR-361-5p in the FOXM1 mRNA. The nucleotide positions are indicated based on the goat FOXM1 mRNA sequence with accession number XM_001314325.1 at NCBI online database (https://www.ncbi.nlm.nih.gov). (E) The construction strategies of FOXM1 mRNA mutant (FOXM1 mRNA-MUT) for the miR-361-5p binding sites. (F) MiR-361-5p directly binds with FOXM1 mRNA. (G) Overexpression of miR-361-5p led to the significant decreasing expression of FOXM1 mRNA in SHF-stem cells. (H) Knockdown of miR-361-5p led to the significant increasing expression of FOXM1 mRNA in SHF-stem cells. The ‘**’ and ‘***’ stand for indicating significant difference with p < 0.01 and p < 0.001, respectively.
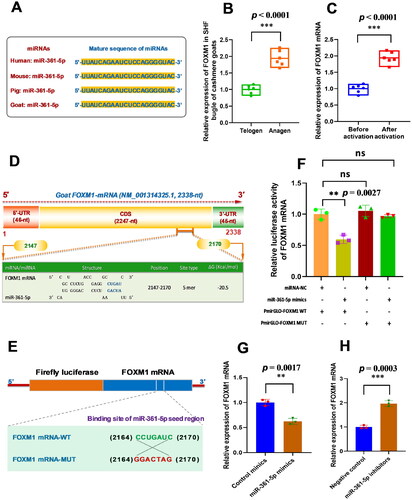
Figure 3. Effects of FOXM1 on the expression of wnt/β-catenin pathway related genes in SHF-stem cells. (A) Overexpression efficiency analysis of FOXM1 in SHF-stem cells of cashmere goats. (B) Konckdown efficiency analysis of FOXM1 in SHF-stem cells of cashmere goats. (C) Overexpression of FOXM1 led to the significant increasing expression of wnt/β-catenin pathway related genes in SHF-stem cells including CTNNB1, LEF1, c-Myc, and CCND1. (D) Knockdown of FOXM1 led to the significant decreasing expression of Wnt/β-catenin pathway related genes in SHF-stem cells including CTNNB1, LEF1, c-Myc, and CCND1. The ‘**’ and ‘***’ stand for indicating significant difference with p < 0.01 and p < 0.001, respectively.
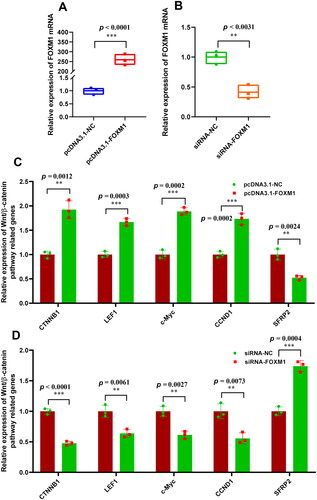
Figure 4. MiR-361-5p negatively regulates the wnt/β-catenin pathway activation through FOXM1 in SHF-stem cells. (A) Overexpression of miR-361-5p led to the significant decreasing expression of wnt/β-catenin pathway related genes in SHF-stem cells including CTNNB1, LEF1, c-Myc, and CCND1. (B) Knockdown of miR-361-5p led to the significant increasing expression of wnt/β-catenin pathway related genes in SHF-stem cells including CTNNB1, LEF1, c-Myc, and CCND1. (C) The overexpression of FOXM1 rescued its mRNA expression level when miR-361-5p mimics was co-transfected in the SHF-stem cells. (D) The knockdown of FOXM1 balanced the up-regulation of FOXM1 mRNA when miR-361-5p inhibitor was co-transfected in SHF-stem cells. (E) The overexpression of miR-361-5p significantly downregulated the activity of β-catenin-driven transcription in the SHF-stem cells with the TOP/FOP flash assay. (F) The knockdown of miR-361-5p significantly upregulated the activity of β-catenin-driven transcription in the SHF-stem cells with the TOP/FOP flash assay. The ‘*’, ‘**’ and ‘***’ stand for indicating significant difference with p < 0.05, p < 0.01 and p < 0.001, respectively.
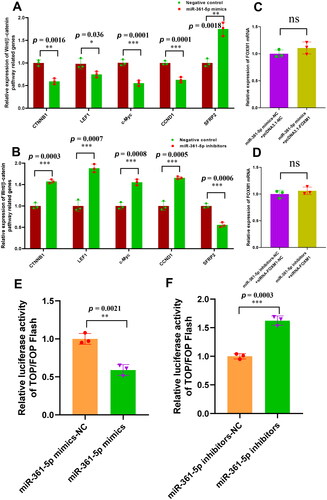
Figure 5. A summary schematic diagram of functional role of miR-361-5p in negatively regulating the induced activation of SHF-stem cells in cashmere goats through FOXM1 mediated wnt/β-catenin pathway.