Figures & data
Figure 1. Structure prediction of the yak AIFM2 protein and its amino acid sequence alignment. (A) Secondary structure prediction of the yak AIFM2 protein. Alpha-helix, beta-turn, extended strand, and random coil are marked in blue, green, red, and yellow, respectively. (B) Tertiary structure prediction of the yak AIFM2 protein. (C) Amino acid sequence alignment of the yak AIFM2 protein and other species. Identical and similar residues are highlighted in yellow and red, respectively.
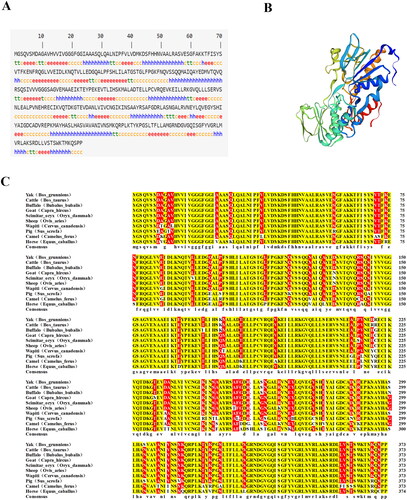
Figure 2. mRNA Expression profile of AIFM2 gene in tissues of yak. The RT-qPCR results were analyzed using the 2−Δct method.Citation26 Different letters upon panels mean significant difference.
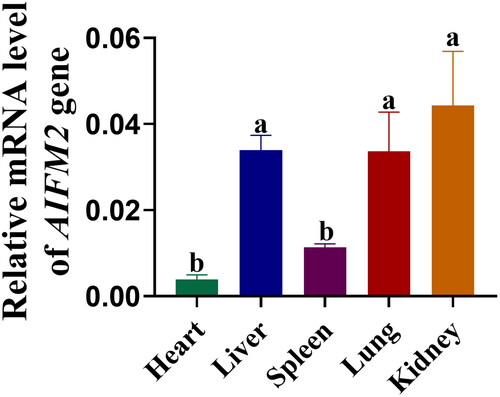
Figure 3. Isolation, culture, and identification of YSFs. (A) Bright-field observation photographs of fibroblasts isolated and cultured using tissue block method. Scale = 500 μm. (B) Bright-field observation photographs of fibroblasts isolated and cultured by enzymatic method. Scale = 500 μm. (C) Immunofluorescence staining of VIMENTIN in enzymatic method-derived YSFs. Scale = 100 μm. (D) Immunofluorescence staining of H3K27me3 in enzymatic method-derived YSFs. Scale = 50 μm.
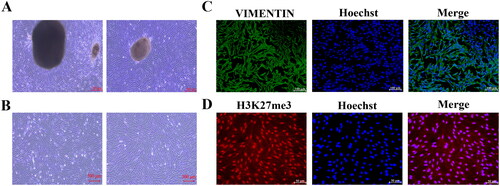
Figure 4. Establishment of BPA-induced toxicity model of YSFs. YSFs were treated with different concentrations of BPA (Ctrl, 100, 150, 200, and 250 μM) for 24 h. (A) Representative images of bright field for YSFs after BPA treatment. Scale = 500 μm. (B) Cell viability of YSFs after BPA treatment. (C) Representative images of reactive oxygen species (ROS) staining for YSFs after BPA treatment. Scale = 100 μm. (D) Ratio of fluorescence intensity statistics (green/blue) corresponding to ROS staining. (E) Content of GSH in YSFs after BPA treatment. (F) Representative images of BODIPY 581/591 C11 staining after BPA treatment. Scale = 50 μm. (G) Ratio of fluorescence intensity statistics (green/red) corresponding to BODIPY 581/591 C11 staining. (H) Content of MDA in YSFs after BPA treatment. Different letters upon panels mean significant difference.
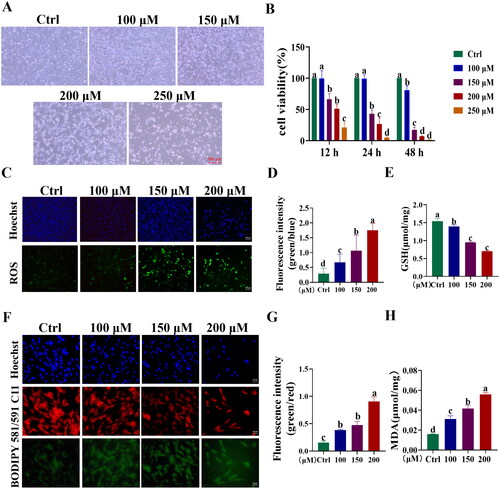
Figure 5. Validation of BPA-induced ferroptosis in YSFs. YSFs were treated with BPA (150 μΜ) and Fer (1, 10, 100 μΜ) for 24 h. (A) Representative images of bright field for YSFs after BPA and Fer treatment. Scale = 500 μm. (B) Cell viability of YSFs after BPA and Fer treatment. (C) Representative image of ROS staining for YSFs after BPA and Fer treatment. Scale =100 μm. (D) Ratio of fluorescence intensity statistics (green/blue) corresponding to ROS staining. (E) GSH content in YSFs after BPA and Fer treatment. (F) Representative images of BODIPY 581/591/C11 staining for YSFs after BPA and Fer treatment. Scale = 50 μm. (G) Ratio of fluorescence intensity statistics (green/red) corresponding to BODIPY 581/591 C11 staining. (H) Content of MDA in YSFs after BPA and Fer treatment. Different letters upon panels mean significant difference.
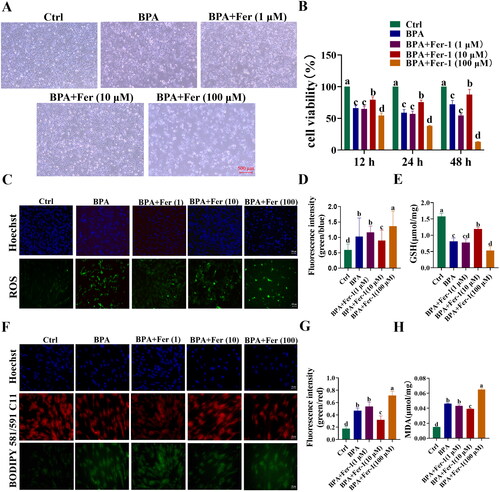
Figure 6. Overexpression of AIFM2 gene alleviates BPA-induced ferroptosis in YSFs. NC: YSFs with pcDNA3.1 (+) transfection. OE: YSFs with pcDNA3.1 (+)-AIFM2 transfection. BPA: YSFs with 150 μM BPA treatment. OE + BPA: YSFs with pcDNA3.1 (+) – AIFM2 transfection and BPA treatment. (A) Construction of pcDNA3.1 (+) – AIFM2 overexpression vector. (B) Relative expression level of AIFM2 in YSFs transfected with pcDNA3.1 (+) – AIFM2. RT-qPCR results were analyzed using the 2−ΔΔct method. (C) Verification of AIFM2 overexpression at protein level. (D) Relative expression level of BAX, BCL-2 and GPX4 in YSFs transfected with pcDNA3.1 (+) – AIFM2. RT-qPCR results were analyzed using the 2−ΔΔct method. (E) Representative photos of bright field for YSFs after pcDNA3.1 (+) – AIFM2 transfection and/or BPA treatment. Scale = 500 μm. (F) Cell viability of YSFs after pcDNA3.1 (+) – AIFM2 transfection and/or BPA treatment. (G) Representative images of reactive oxygen species (ROS) staining for YSFs after pcDNA3.1 (+) – AIFM2 transfection and/or BPA treatment. Scale = 100 μm. (H) Ratio of fluorescence intensity statistics (green/blue) corresponding to ROS staining. (I) Content of GSH in YSFs after pcDNA3.1 (+) – AIFM2 transfection and/or BPA treatment. (J) Representative image of BODIPY 581/591/C11 staining for YSFs after pcDNA3.1 (+) – AIFM2 transfection and/or BPA treatment. Scale = 50 μm. (K) The ratio of fluorescence intensity statistics (green/red) corresponding to BODIPY 581/591 C11 staining. (L) Content of MDA in YSFs after pcDNA3.1 (+) – AIFM2 transfection and/or BPA treatment. Different letters upon panels mean significant difference.
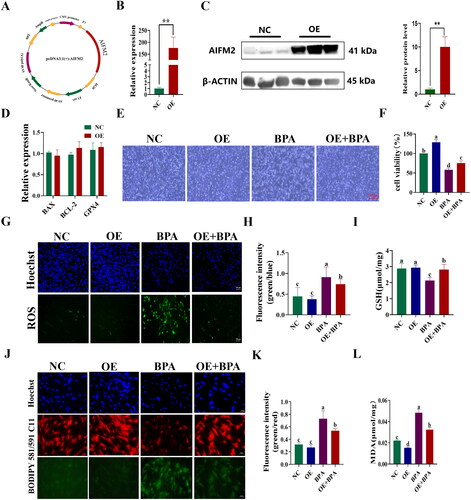
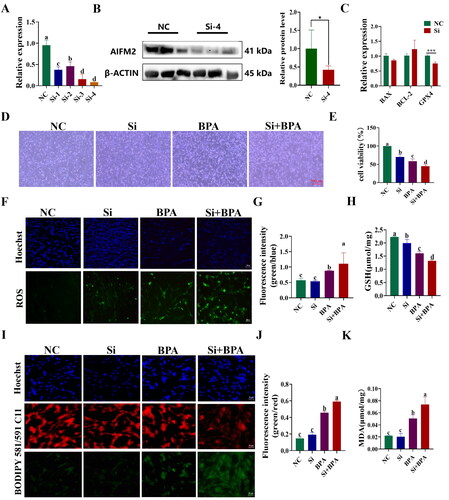
Figure 7. Interference with AIFM2 gene exacerbates BPA induced ferroptosis in YSFs. NC: YSFs with Non-targeted negative control transfection. Si: YSFs with siRNA-4 transfection. BPA: YSFs with 150 μM BPA treatment. Si + BPA: YSFs with siRNA-4 transfection and BPA treatment. (A) Relative expression level of AIFM2 after specific siRNAs interference in YSFs. The RT-qPCR results were analyzed using 2−ΔΔCt method. (B) Validation of AIFM2 inteference at protein level. (C) Relative expression level of BAX, BCL-2 and GPX4 in YSFs transfected with siRNA-4. RT-qPCR results were analyzed using the 2−ΔΔct method. (D) Representative photos of bright field for YSFs after siRNA-4 transfection and/or BPA treatment. Scale = 500 μm. (E) Cell viability of YSFs after siRNA-4 transfection and/or BPA treatment. (F) Representative images of ROS for YSFs after siRNA-4 transfection and/or BPA treatment. Scale = 100 μm. (G) Ratio of fluorescence intensity statistics (green/blue) corresponding to ROS staining. (H) Content of GSH in YSFs after siRNA-4 transfection and/or BPA treatment. (I) Representative image of BODIPY 581/591/C11 staining for YSFs after siRNA-4 transfection and/or BPA treatment. Scale = 50 μm. (J) Ratio of fluorescence intensity statistics (green/red) corresponding to BODIPY 581/591 C11 staining. (K) Content of MDA in YSFs after siRNA-4 transfection and/or BPA treatment. Different letters upon panels mean significant difference.