Figures & data
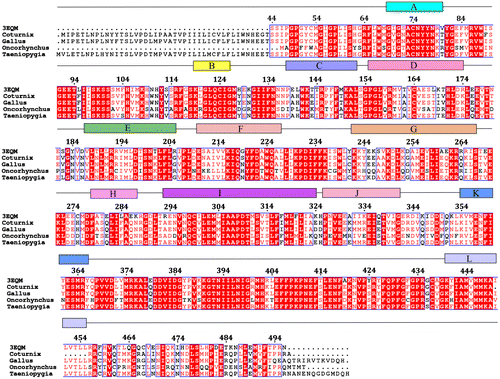
Figure 2. (a) The alignment of homology-modelled proteins of C. japonica, G. gallus, T. guttata and O. mykiss with human protein. (b) Comparison of the important residues at the catalytic site of the aromatase enzyme in the studied species. In the three bird species, V373 in humans is replaced by I373, while in O. mykiss it is replaced by T373. L372 in human is replaced by F372 in O. mykiss.
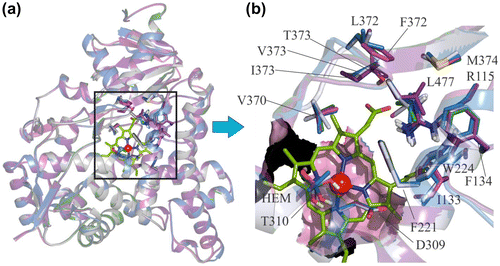
Figure 4. (a) Docking of fadrozole (S), (b) imazalil (S) and (c) vorozole (S) at the active site of human aromatase.
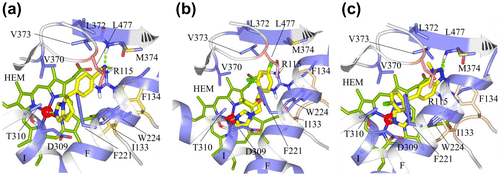
Figure 5. Docking of fadrozole (S), imazalil (S) and vorozole (S) at the C. japonica aromatase (a–c), at the G. gallus aromatase (d–f), at the T. guttata aromatase (g–i), and at the O. mykiss aromatase (j–l).
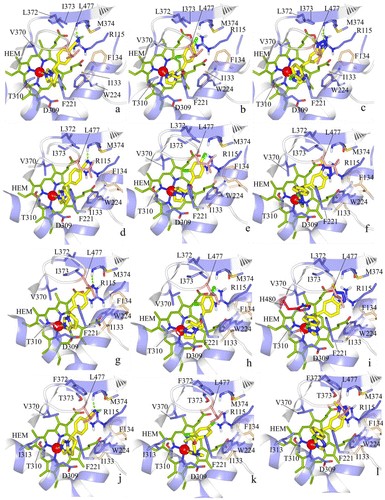
Table 1. Interactions of fadrozole, imazalil and vorozole with the active site residues of human, bird, and fish aromatases.
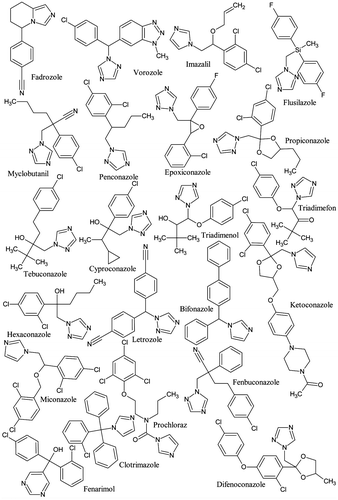
Table 2. Docking scores (Kcal/mol) vs. inhibition of human recombinant CYP19 (aromatase) for 18 azoles.
Table 3. Docking scores (Kcal/mol) of azoles in the aromatases of the different studied species.
Figure 6. Correlation between the docking scores of 18 azoles on the human aromatase and the corresponding human recombinant CYP19 inhibitory activity values (Table ).
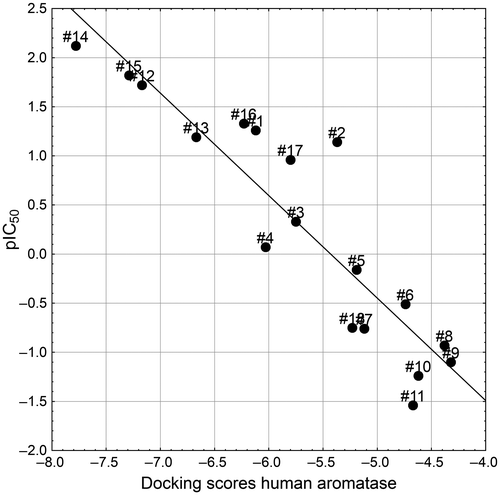
Table 4. Docking scores (Kcal/mol) and inhibition of aromatase activities in brain and ovarian microsomes of O. mykiss [Citation28].
Figure 7. Differences in the residuals obtained for the 10 brain pIC50 values in O. mykiss (Table ).
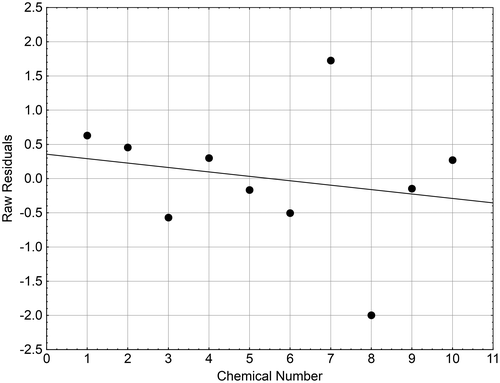
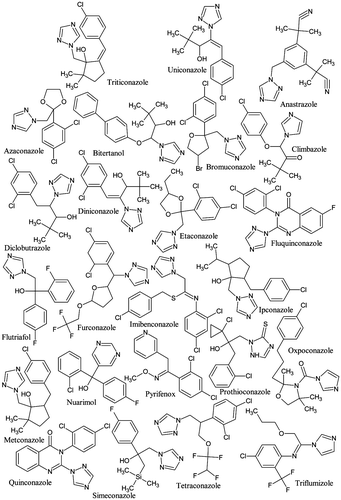
Table 5. Docking scores (Kcal/mol) of 24 azoles on aromatases of the different studied species.
Figure 9. Correlation matrix of the score values on the aromatases of the different studied species for the 24 compounds in Figure .
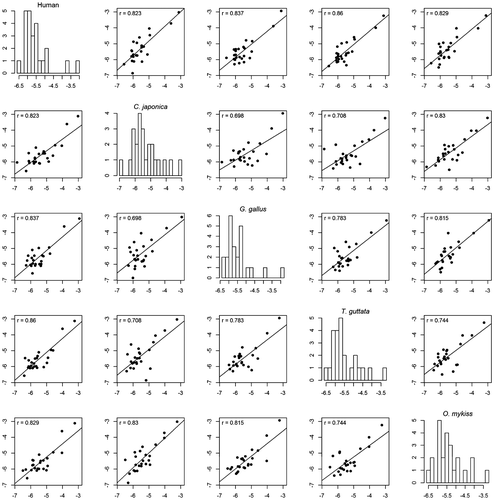
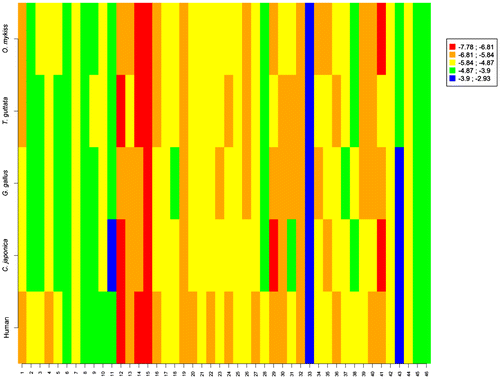
Figure 10. Graphical display of the score values obtained for the 46 studied molecules (x-axis) on the aromatases of human, C. japonica, G. gallus, T. guttata and O. mykiss.