Figures & data
Figure 1. Synthesis of a conjugate between a cell-penetrating peptide (CPP) and a cationic oligolysine peptide (CP) via a disulfide linkage.
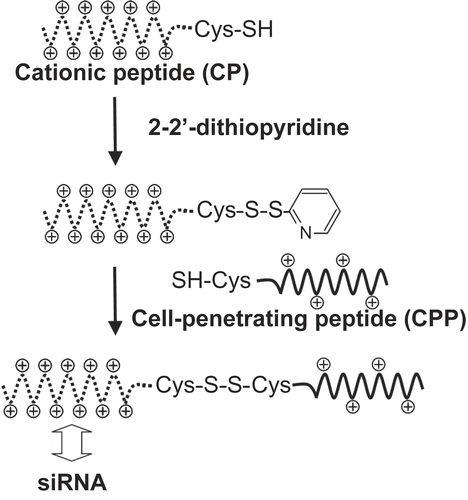
Figure 2. Formation of complexes between TP-CP conjugates and siRNA. Various amounts of TP-CP conjugates and a constant amount of fluorescein-labeled siRNA were mixed in phosphate buffer ( 10 mm, pH 7.2). Migration of siRNA was analyzed by 20% PAGE and fluorescence images of gels were shown.
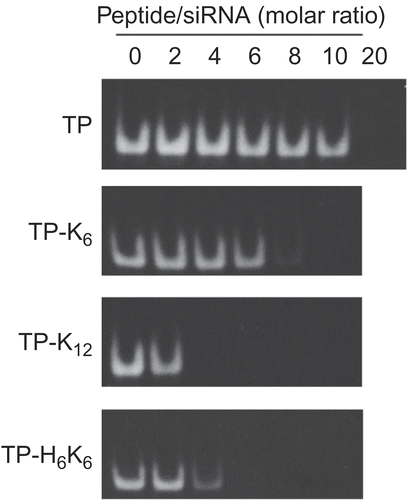
Table 1. Cytotoxicity of complexes between peptide conjugates and siRNA.
Figure 3. Internalization of siRNA complexes formed with various peptide conjugates by FRSK cells. The cells were incubated for 2 hr at 37°C with mixtures of fluorescein-labeled siRNA (final concentration = 60 nm) and various peptides at a 20-fold (open bars) or 60-fold (closed bars) molar ratio (peptide/siRNA). Then cells were recovered by trypsinization and the fluorescence intensity of the cell lysate was measured. Results were normalized by the amount of protein in the cell lysate. Each data point represents the mean ± SD of three wells.
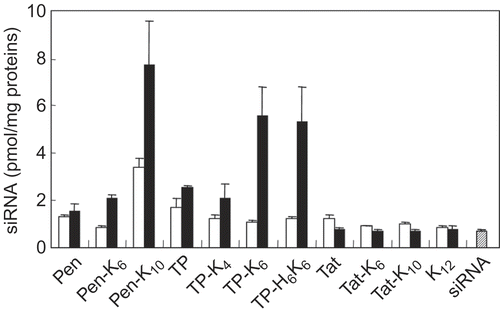
Figure 4. Influence of various endocytosis inhibitors on the uptake of siRNA with peptide conjugates by FRSK cells. The cells were pre-incubated in the presence of each endocytosis inhibitor before addition of the complexes. Then cells were incubated for 2 hr at 37°C with a mixture of fluorescein-labeled siRNA (final concentration = 60 nm) and Pen-K10 (open bars) or TP-K6 (closed bars) at a 60-fold molar ratio (peptide/siRNA). Each data point represents the mean ± SD of three wells. Asterisks (* p < 0.05, ** p < 0.01) indicate values that significantly differ from those measured for cells treated without inhibitors.
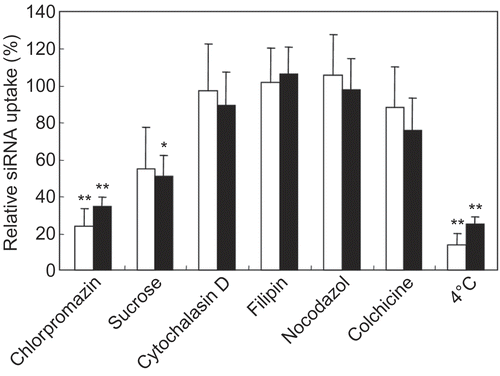
Figure 5. Fluorescence images of living FRSK cells after treatment with complexes of peptide conjugates and siRNA. A mixture of fluorescein-labeled siRNA (final concentration = 60 nm) with (A) Pen-K10 or (B) TP-K6 at a 60-fold molar ratio (peptide/siRNA) was incubated with the cells for 2 hr at 37°C, after which fluorescence microscopy was done without fixation of the cells. (C) LF 2000 was also tested as a siRNA carrier. Scale bar = 50 μm.
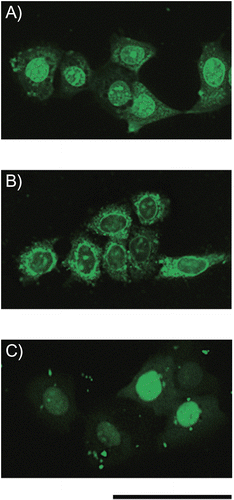
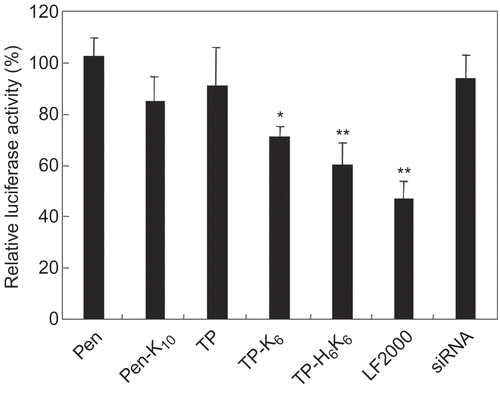
Figure 6. Luciferase gene silencing by siRNA with peptide conjugates. Mixtures of siRNA (anti-luciferase or negative control siRNA) with various peptide conjugates at a 60-fold molar ratio (peptide/siRNA) were incubated with the cells for 2 hr at 37°C. The final siRNA concentration was 60 nm. Subsequently, the cells were incubated with complexes of LF2000 and pCMV-luciferase plasmid, after which luciferase expression was determined by using a Luciferase Assay System. Silencing of luciferase gene expression is shown as the relative ratio of luciferase activity induced by different siRNA (anti- luciferase siRNA/negative control siRNA). Each data point represents the mean ± SD of three wells. Asterisks (* p < 0.05, ** p < 0.01) indicate values that significantly differ from those measured for cells treated with siRNA.