Figures & data
Figure 1 The SE-HPLC profile of the native HSA to detect the elution times of the native HSA, HSA-HSA dimer, and HSA aggregate. The real line and the dashed line showed the adsorbance at 280 nm and 405 nm, respectively.
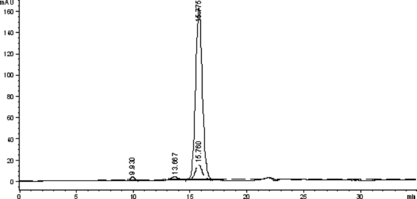
Figure 2 The SE-HPLC profile of the native Hb to detect the elution times of the native Hb and Hb-Hb dimer. The real line and the dashed line showed the absorbance at 280 nm and 405 nm, respectively.
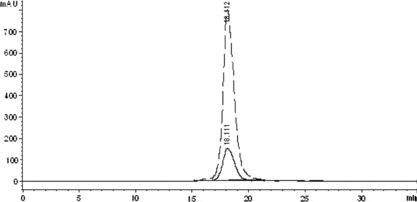
Figure 3 The SE-HPLC profile of the Hb treated by glutaraldehyde to detect the elution time of the intra-crosslinked Hb and inter-crosslinked Hb-Hb dimer. The real line and the dashed line showed the adsorbance at 280 nm and 405 nm, respectively.
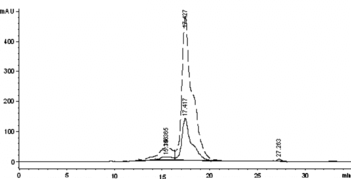
Figure 4 Characterization of the HSA-HSA dimer by SDS-PAGE. Lane 1 was crosslinked HSA treated by glutaraldehyde, Lane 2 was native HSA, and lane 3 was standard marker.
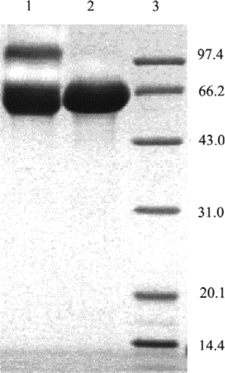
Figure 5 Characterization of the covalently crosslinked subunits of Hb by SDS-PAGE. Lane 1: standard marker; Lane 2: native Hb; and lane 3: Hb sample treated with glutaraldehyde.
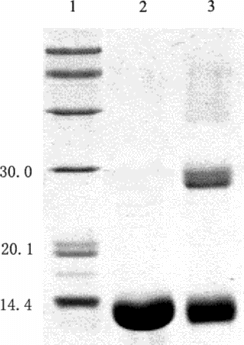
Figure 6 The effect of pH value on the content of conjugate or byproducts in the conjugation reaction. The conjugation reaction was carried out at 4°C for 1 h under different pH values, and the reaction was stopped by addition of excessive lysine solution. The components were determined by TSK G3000 column. The yields of the byproducts and target 1:1 Hb-HSA conjugate were estimated by integrating the chromatograms of the reaction products. The results were obtained from the average of three repeated measurements.
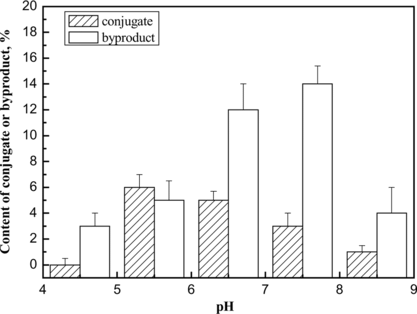
Figure 7 Effect of concentration and molecular weight of PEG on the conjugation reaction of Hb and HSA. The optimum condition is annotated by arrow.
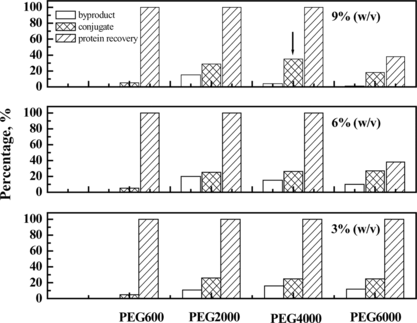
Table 1. Summary of the molecular weight parameters of the peaks corresponding to detected by MALLS
Figure 8 The profiles of the reaction mixture prepared in presence of PEG (A) and in absence of PEG (B), respectively. The samples were loaded on a size-exclusion high performance TSK-GEL G3000SW column, followed by elution at a flow rate of 0.5 ml min-1 50 mM PBS containing 0.1 M Na2SO4.
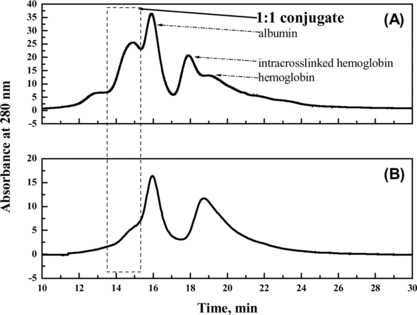
Figure 9 Characterization of the reaction mixture using SDS-PAGE. Lane1: native hemoglobin; lane 2: native albumin; lane 3: standard marker; lane 4: reaction mixture in the presence of PEG; lane 5: reaction mixture in the absence of PEG. All samples were boiled for 5 min. The stacking and running gels were 5 and 12% in acrylamide, respectively. The numbers on the left side of the gel indicated molecular weight of marker.
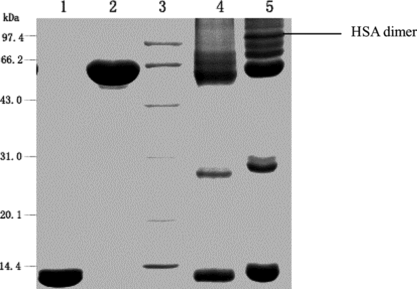
Figure 10 SE-HPLC and MALLS analyses of reaction mixture obtained under crowding condition by PEG. LS was laser signal; AUX1 was refractive index; AUX2 was absorption at 280 nm. The MALLS instrument was calibrated by standard monomer HAS. Astra V.4.9 software was used to calculate the molecular weights (the results are summarized in ). The flow rate was 0.4 ml/min. Peak 1 was the byproduct; Peak 2 was the target conjugate of 1:1 Hb-HSA; Peak 3 and peak 4 were the unreacted HSA and unreacted Hb, respectively.
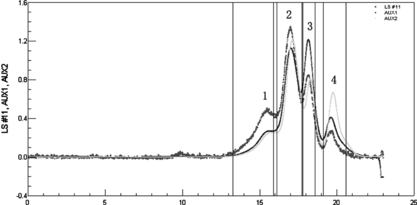
Table 2. Comparison of bioactivity of conjugates prepared under crowding and non-crowding conditions
Figure 11 O2 equilibrium curves of native bovine hemoglobin and the conjugates prepared under crowding (created by adding PEG with optimum molecular weight and concentration) and non-crowding conditions.
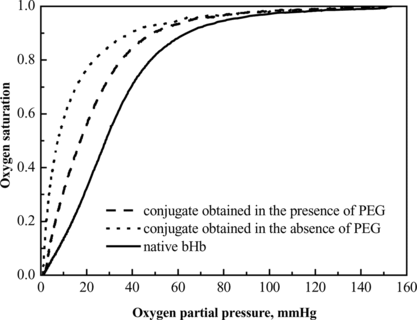
Figure 12 Effect of protein concentration on the content of conjugate or byproducts in the conjugation preparation obtained in the absence of PEG, compared with the conjugation product prepared in the presence of PEG at the apparent protein concentration of 2 mg/ml, to investigate the effect of PEG on the conjugation reaction.
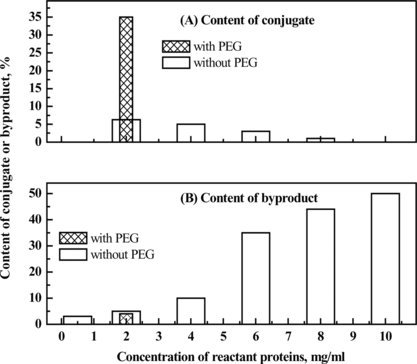
Figure 13 Isoelectric focusing of Hb and/or HSA treated with glutaraldehyde. Lane 1: HSA reacted with glutaraldehyde; Lane 2: Hb reacted with glutaraldehyde; Lane 3: the resulting mixture of the conjugation reaction; Lane 4: standard marker; Lane 5: native Hb; Lane 6: native HSA. Employing conditions described in Materials and Methods.
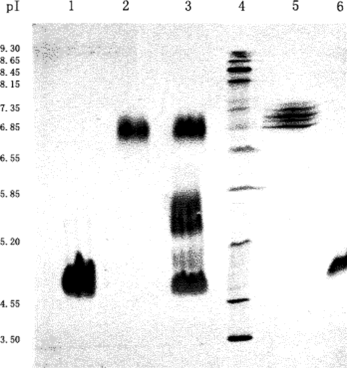
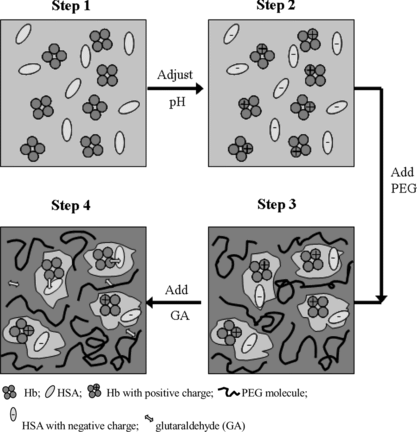
Figure 14 A scheme of the presumable conjugation process assisted by PEG. Step 1: Hb and HSA were dissolved; Step 2: the pH value was adjusted to pH 5.7 (the average of pI of Hb and that of HSA), and Hb molecule was positively charged and HSA was negatively charged; Step 3: PEG was added to adjust the physical environment of Hb and HSA. The oppositely charged Hb and HSA were assumed to form an electrostatic-interacting pair, and micro-compartmented into one pore in the network of PEG with proper concentration and molecular weight; Step 4: Glutaraldehyde was added, and conjugation of Hb and HSA in 1:1 ratio occurred, which simultaneously lost its net charge and then might further couple to the byproduct comprising Hb and HSA in 2:2 ratio (dimer of the target conjugate).