Figures & data
Figure 1. Schematic representation of peptide-PEG-Hb synthesis. Peptide in HEPES buffer (5 mM HEPES, pH 7) reacted to MAL-PEG-NHS for 3 min at room temperature. The molar ratio is 1:1 in anaerobic conditions. Using a 1.2:1 molar ratio of peptide-PEG solution to deoxyhemoglobin in HEPES buffer (8.3 g/L NaCl and 20 mM HEPES, pH 8) was incubated in room temperature for 1 h. MAL and NHS are the functional groups in PEG for crosslinking with cysteine in the peptide and lysine in the outer surface of hemoglobin, respectively.
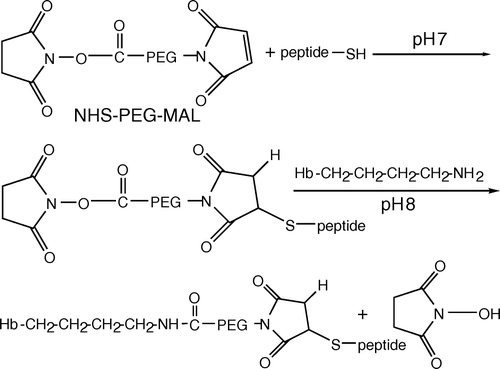
Figure 2. CM-sepharose elution profile and SDS-polyacrylamide gel electrophoresis of the peptide-PEG-Hb mixture. (A) Peptide-PEG-Hb was separated by ion exchange chromatography on a CM-sepharose column and monitored at 542 nm. The CM-sepharose column was eluted with each 1 mL, 100–190 mM NaCl difference 10 mM gradient, and later was eluted with 15 mL, 200 mM NaCl and collected 0.5 mL in each fraction. The fraction 10–13, fraction 18–21, and fraction 26–28 were collected as peak1, peak2, and peak3, respectively. (B) 15% SDS-polyacrylamide gel was used to characterized various Hbs. Hb: unmodified Hb; m-Hb: unpurified peptide-PEG-Hb; peak1-3: same as A.
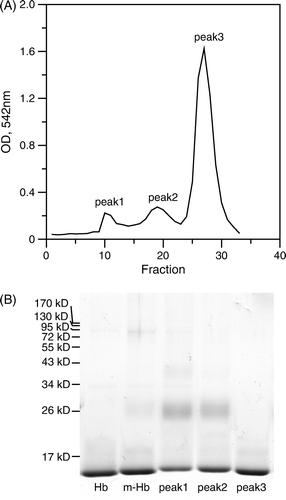
Figure 3. MALDI-TOF mass spectra of various Hbs. Various Hbs were dialyzed against deionized water 3 times to remove salt before analyzing by a MALDI-TOF mass spectrometer. (A): unmodified Hb; (B): peptide-PEG-Hb-1; (C): peptide-PEG-Hb-2.
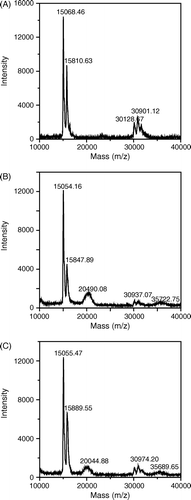
Figure 4. Two dimensional gel electrophoresis of various Hbs. The first dimensional was carried out in 7 cm, pH 3-10 linear gradient IEF gel and the second dimensional was performed using 15% polyacrylamide gels and followed by staining with the Coomassie brilliant blue R-250. (A): unmodified Hb; (B): peptide-PEG-Hb-1; (C): peptide-PEG-Hb-2.
Figure 5. LC-MS/MS chromatographic profiles of the digestion products of various Hbs using TPCK-treated trypsin. (A): unmodified Hb; (B): peptide-PEG-Hb-1; (C): peptide-PEG-Hb-2. The peptide sequences for the corresponding peaks are listed in .
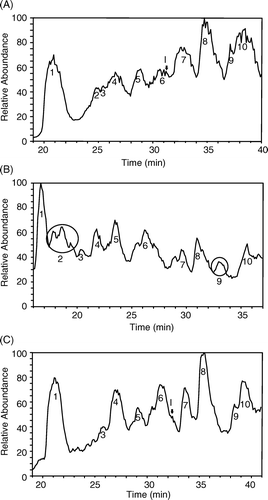
Table 1. The peptide sequences of the chromatographic peaks in LC profiles of trypsinized Hbs
Figure 6. SDS-polyacrylamide gel of polymerization products of peptide-PEG-Hb with SPA-PEG-SPA. Peptide-PEG-Hb and unmodified Hb was mixed in a molar ratio of 1:1 for 1 h. SPA-PEG-SPA in HEPES buffer (5 mM HEPES, pH 7) were then added to the Hb mixture in a molar ratio of 8:1. SPA-1 and SPA-2: polymerization of peptide-PEG-Hb-1/Hb and peptide-PEG-Hb-2/Hb with SPA-PEG-SPA, respectively.
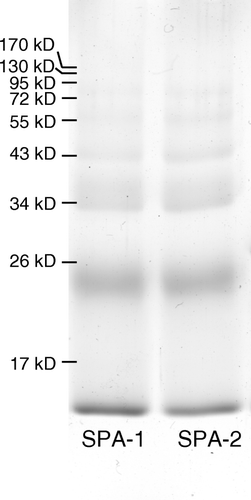
Figure 7. Oxygen dissociation curves for various Hbs. The oxygen dissociation curve was obtained using the PCA (1.4 mM)/PCD (0.06 units/ml) enzyme system. 10 M (in heme) of various Hbs was dissolved in the buffer containing 8.3 g/L NaCl, 20 mM HEPES pH 7.4, and 1 mM EDTA. The data were analyzed using SVD algorithm against oxy-, deoxy-, and metHb standard spectra. (A) unmodified Hb (squares), peptide-PEG-Hb-1/Hb (molar ratio 1:1) (circles), SPA-peptide-PEG-Hb-1 (triangles), and unmodified Hb + 5 mM 2,3-DPG (inverse triangles). The inset shows unmodified Hb (squares), peptide-PEG-Hb-2/Hb (molar ratio 1:1) (circles), SPA-peptide-PEG-Hb-2 (triangles). (B) unmodified Hb (squares), peptide-PEG-Hb-1/Hb (molar ratio 1:1) (circles), cysteine-PEG-Hb/Hb (molar ratio 1:1) (triangles).
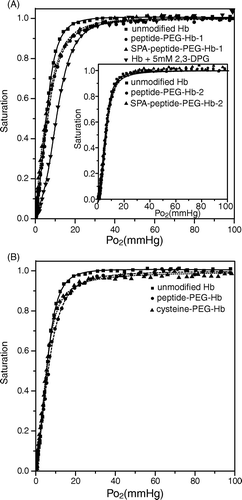
Table 2. Hill coefficients of various Hbs
Figure 8. The distances of possible conjugation locations of the peptide to the 2,3-DPG binding site. This graph and measurements were generated using The PyMOL Molecular Graphics System Citation[22] based on the deoxyHb structure (3HHB).
![Figure 8. The distances of possible conjugation locations of the peptide to the 2,3-DPG binding site. This graph and measurements were generated using The PyMOL Molecular Graphics System Citation[22] based on the deoxyHb structure (3HHB).](/cms/asset/cb6bb8de-5b14-4e15-9332-7611c13e8f60/ianb19_a_366638_f0008_b.gif)