Figures & data
Covariance parameter estimates.
ANOVA table: 1-way.
Figure 1. a, b, and c: QQ-plots of synthetic test data demonstrating that the method constructs lognormal distributions with random selection of 100 values for chosen geometric means (GM = 10) and different geometric standard deviations (GSD); a) GSD = 2; b) GSD = 2.8; and c) GSD = 3.6.
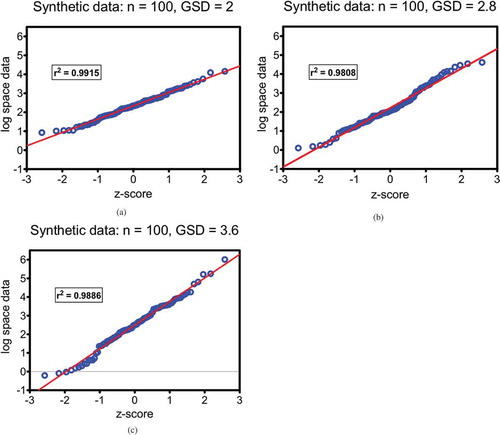
Figure 2. a, b: Comparisons of ICC calculation methods of perfectly balanced data with respect to different data parameters of GSD and target ICC. Each dataset has 100 subjects; a) comparisons for two measurements per subject and b) comparisons for 10 measurements per subject. Differences between Figure 2a and 2b are due to the number repeat measures and not due to the method of calculation. Each test was performed three times.
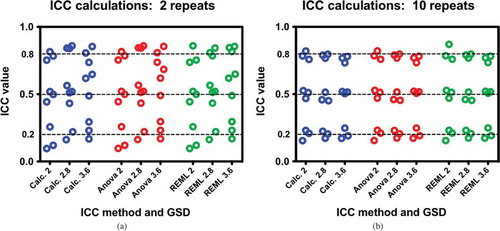
Figure 3. a, b: Comparisons of ICC calculation methods of data missing 10% of the values at random with respect to different data parameters of GSD and target ICC. Each dataset has 100 subjects; a) comparisons for two measurements per subject and b) comparisons for 10 measurements per subject. Differences between Figure 3a and 3b are due to the number repeat measures and not due to the method of calculation. Furthermore, missing at random values do not affect the character of the comparisons of the ICCs. Each test was performed three times.
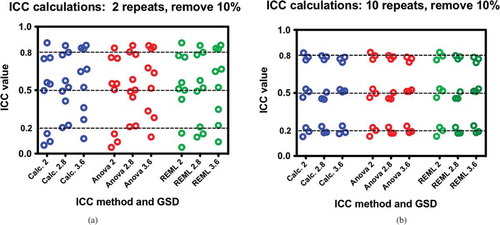
Figure 4. Comparisons of ICC calculation methods with respect to the number of subjects. All data sets have three measures per subject with10% of data removed at random. Scatter increases as number of samples decreases; however, the method of calculation does not affect the results. Each test was performed three times.
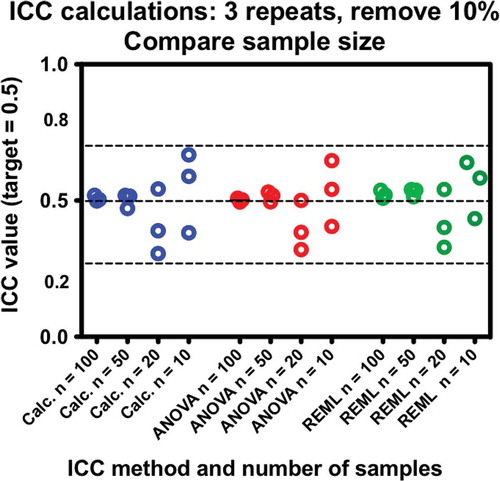
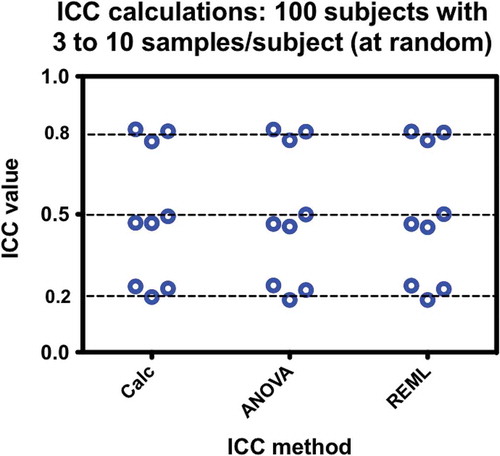
Figure 5. Comparisons of ICC calculation methods with respect to severely unbalanced data. At random, 100 subjects have anywhere from 3 to 10 measurements each for datasets targeted for 0.2, 0.5, and 0.8 ICC values. The method of calculation does not affect the results. Each test was performed three times.