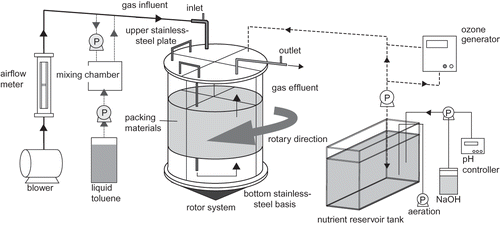
Figures & data
Table 1. Operating conditions of toluene removal experiment using RSB biofilter
Figure 2. Change of parameters during the operation period of the RSB: (a) Toluene removal efficiency (%) and the toluene inlet loading (g m−3 hr−1). (b) Concentration of NH4-N (mg L−1) and SS (mg L−1) in the nutrient tank. Toluene removal efficiency (%) was calculated as follows: (1−Co/Ci) × 100, where Co is the toluene outlet concentration (ppmv); Ci, the toluene inlet concentration (ppmv).
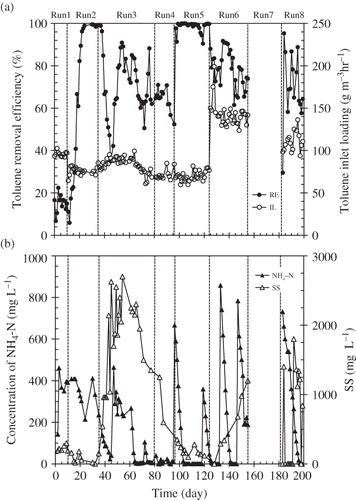
Figure 3. DGGE profiles of universal 16S rRNA gene amplicons collected from the nutrient reservoir tank (no.1–14) and biofilm on the packing material (no.15) of RSB. Open circles with numbers indicate excised bands. These bands were analyzed by sequencing. A phylogenetic tree based on the sequence data is shown in . Lanes: 1; day 2, 2; day 9, 3; day 16, 4; day 31, 5; day 36, 6; day 40, 7; day 51, 8; day 68, 9; day 105, 10; day 118, 11; day 132, 12; day 145, 13; day 163, 14; day 183, M; marker, 15; day 68.
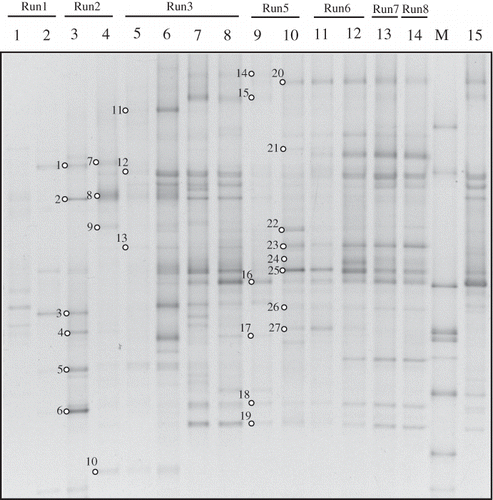
Figure 4. Nonmetric multidimensional scaling analysis of the DGGE fingerprint (stress value is 0.04). Numbers 1–15 indicate sample numbers from the DGGE analysis (see and ). Numbers 1–14 (solid circles) were samples of the nutrient solution, and number 15 (open circle) was a sample of the biofilm on the packing material.
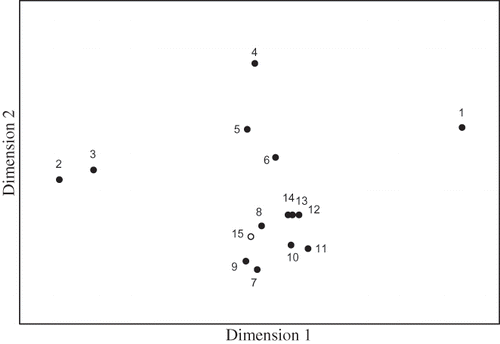
Figure 5. DGGE profile of tmoA-like gene amplicons collected from the nutrient reservoir tank of RSB. Arrows indicate the excised band.
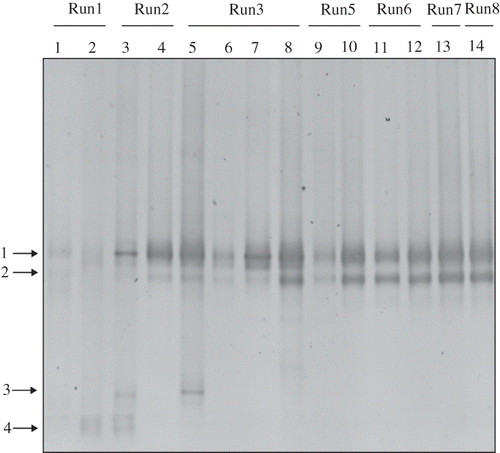
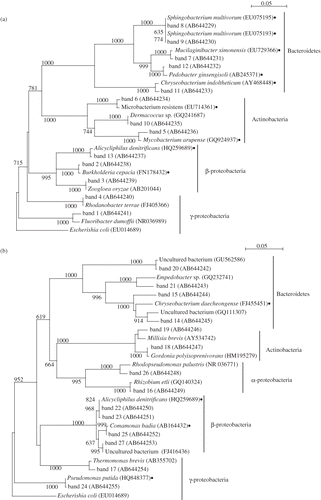
Figure 6. Neighbor-joining tree of partial 16S rRNA gene sequence (∼550 bp) recovered by denaturing gradient gel electrophoresis bands. Accession numbers appear after the genus name. The neighbor-joining tree was constructed as described in the text. Each sequence, except for those from this study, was obtained from the DDBJ (DNA Data Bank of Japan). The scale bar indicates 0.05 changes per nucleotide. The numbers on the branches refer to bootstrap values for 1000 replicates; only those above 600 are shown. Solid circles mean the reported bacteria that can utilize or tolerate to the toluene in the papers.