Figures & data
Table 1. Primer sets for real-time RT-PCR used in this study.
Figure 1. CircRNA_0044556 was highly expressed in TNBC, while miR-665 was low expressed. (A) The level of circRNA_0044556 in cells was analyzed by RT–qPCR. (B) The level of miR-665 in cells was analyzed by RT–qPCR. **p < .01, ***p < .001 vs. MCF-10A, N = 3. *p < .05, ***p < .001 vs. MDA-MB-231, N = 3.
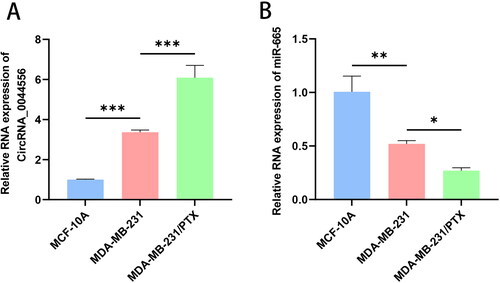
Figure 2. Down-regulated circRNA_0044556 inhibited the malignant phenotype and paclitaxel resistance of TNBC cells. (A) RT–qPCR was used to assess the endogenous expression of circRNA_0044556 after transfection of cells. (B) MTT assays were used to assess cell proliferation. (C) Clonal formation assays were used to assess cell proliferation. (D) Apoptosis was measured by FCM. (E–H): WB analysis (E) of the expression levels of Bax (F), Bcl-2 (G) and Ki67 (H) proteins. **p < .01, ***p < .001 vs. sh-NC, N = 3.
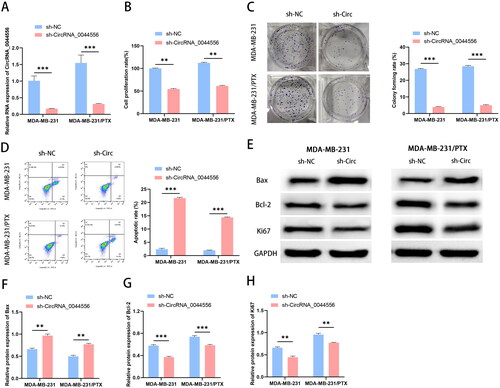
Figure 3. CircRNA_0044556 competitively adsorbs miR-665. (A) Predicted binding sites of circRNA_0044556 and miR-665. (B) Dual luciferase reporting experiment verified the targeting relationship between circRNA_0044556 and miR-665. (C) The expression of miR-665 in MDA-MB-231 cells with circRNA_0044556 knockdown or overexpression was analyzed by RT–qPCR. **p < .01, vs. NC mimics, N = 3. ***p < .001, vs. sh-NC, N = 3. **p < .01, vs. pcDNA 3.1, N = 3.
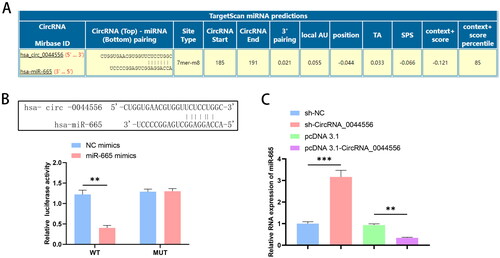
Figure 4. CircRNA_0044556 affects the malignant phenotype and paclitaxel resistance of TNBC cells by regulating miR-665 expression. (A) RT–qPCR was used to assess the endogenous expression of circRNA_0044556 after transfection of cells. (B) MTT assays were used to assess cell proliferation. (C) Clonal formation assays were used to assess cell proliferation. (D) Apoptosis was measured by FCM. (E–H) WB analysis (E) of the expression levels of Bax (F), Bcl-2 (G) and Ki67 (H) proteins. *p < .05, **p < .01, ***p < .001 vs. sh-circRNA_0044556 + in-NC, N = 3.
Supplemental Material
Download MS Word (1.4 MB)Data availability statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
