Figures & data
Figure 1 a, The haploid chromosome set of an oocyte I from a hybridogenetic R. esculenta specimen of the L–E system: the 13 univalents are identifiable as belonging to R. ridibunda lampbrush genome on the basis of specific cytogenetic markers: centromeres are always evident as loopless granules or bars and a few giant lateral structures are inserted along the chromosomes. The haploid set of 13 ridibunda univalents can be explained by assuming that this oocyte has excluded the lessonae genome without accomplishing the subsequent endoreduplication. b,c, Chromosome preparations of germinal tissues hybridised in situ with DIG‐labelled RrS1 as a probe. Spermatogonial metaphases (male specimen, cross 02.41) containing either 13 ridibunda chromosomes, 6 of which show RrS1‐marked centromeres (b) or 26 ridibunda chromosomes, 12 of which have RrS1‐marked centromeres (c). These findings reflect germ cells after the elimination of lessonae genome, but prior to the endoreduplication of the remaining ridibunda genome (b), and both the exclusion and subsequent endoreduplication of the ridibunda genome, respectively (c). Phase contrast (a); c = centromeres; S = sphere. Scale bar: 20 µm (a), 5 µm (b, c).
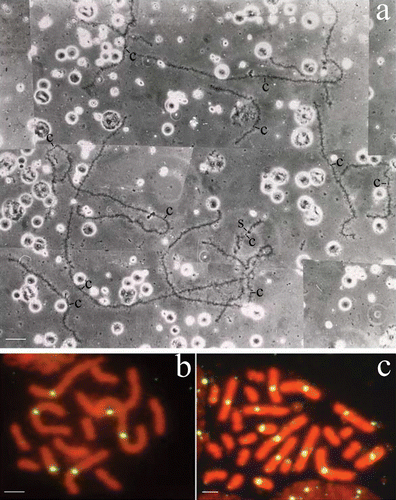
Figure 2 Expression profile ofRlVlg (a), RlPL10 (b) and RlYB2 (c) during oogenesis of R. ridibunda (a), R. lessonae (b), and R. esculenta (c), by using whole‐mount (a, c) and on section (b) in situ hybridisations. Oocyte stages are indicated. Scale bars: 100 µm.
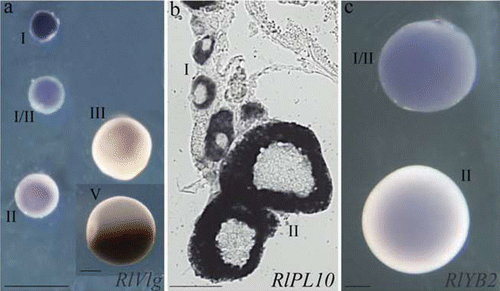
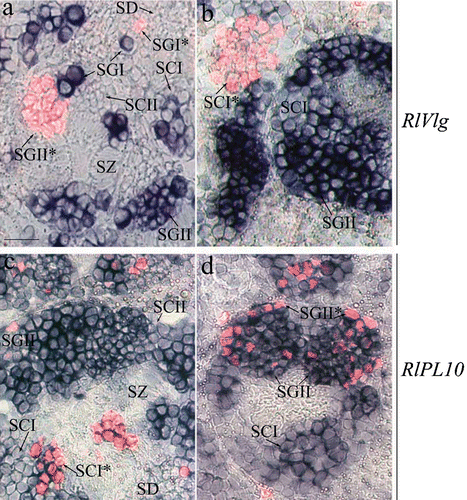
Figure 3 Expression pattern ofRlVlg (a, b) and RlPL10 (c, d) during spermatogenesis. Hybridised sections of adult testes of R. ridibunda (a, c) and R. esculenta (b, d) were immunostained with anti‐phosphohistone H3 antibody. RlVlg (blue) and phosphohistone H3 (red fluorescence) expression signals were not overlapping. The RlPL10 expression pattern (blue‐marked) included phosphohistone H3‐positive cells. SGI = primary spermatogonia, SGII = secondary spermatogonia, SCI = primary spermatocites, SCII = secondary spermatocytes, SD = spermatids, SZ = spermatozoa. Scale bar: 40 µm.