Figures & data
Table I. Genetic diversity estimates of C. dilatata and C. fecunda. N refers to the number of individuals sampled, S is the number of segregating sites, H is the number of haplotypes, and for all populations refers to unique haplotypes. Hd is the haplotype diversity and π is the nucleotidic diversity, both with standard deviation in parenthesis. *Significant values at p < 0.05. NA, not applicable
Figure 1. Map of the Chilean and Argentinean coasts showing the sampling locations of animals used in this study. 1, Coquimbo; 2, Corral; 3, Chinquihue; 4, Metri; 5, Quempillén; 6, Aucho; 7, Yaldad; 8, Melinka; 9, Puerto Madryn. Ancon and Santa María locations from Perú are not shown, as such sequences were obtained from GenBank.
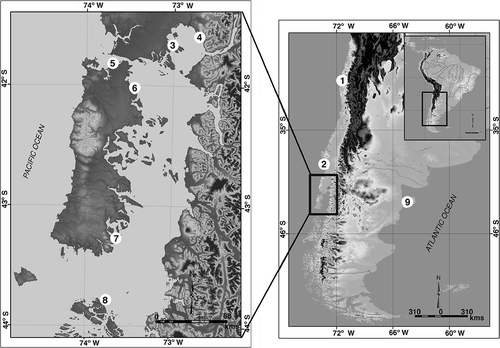
Table II. Distribution of haplotypes per locality of C. dilatata.
Table III. Distribution of haplotypes per locality of C. fecunda
Figure 2. Bayesian tree representing the relationships between C. fecunda and C. dilatata populations. Numbers above the branches represent bootstrap support value and Bayesian posterior probabilities. Numbers in parenthesis indicate the number of individuals sharing the respective sequence.
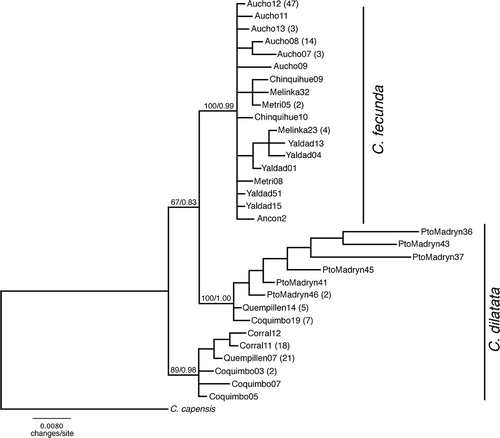
Figure 3. Median-joining network representing the relationships among haplotypes of Crepipatella dilatata (A) and Crepipatella fecunda (B). Small black circles indicate median vectors (Bandelt et al. Citation1999). Circle size is proportional to haplotype frequencies, with the smaller circle representing one sample. Numbers at the branches represent the number of mutations separating the haplotypes if greater than one.
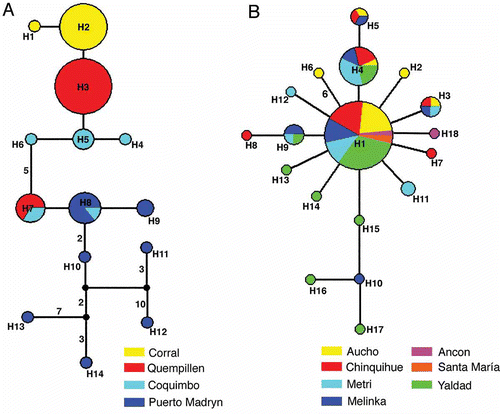
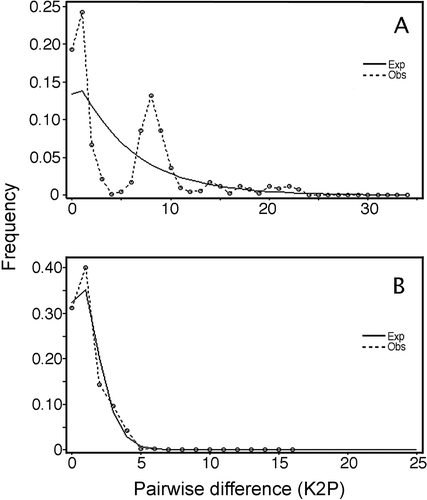
Figure 4. Mismatch distributions observed (Obs) for (A) Crepipatella dilatata and (B) Crepipatella fecunda. The expected (Exp) frequency is based on a sudden population expansion model. Goodness-of-fit of the observed versus the theoretical mismatch distribution was tested using the sum of squared deviations (SSD).