Figures & data
Figure 1. (a) Location of the area of interest in Argentina (white square) and (b) study areas in the four invasion foci located next to the cities (black dots) of Luján and Escobar in Buenos Aires province, Cañada de Gómez in Santa Fé province, and La Cumbrecita in Córdoba province (lines divide provinces).
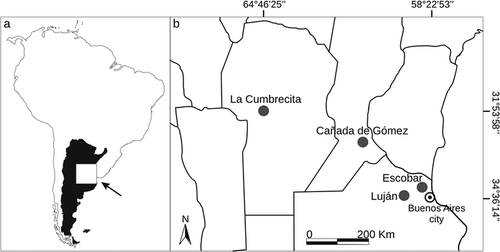
Figure 2. Bayesian inference trees of Callosciurus genus. Tree derived from D-loop. Numbers next to branches are Bayesian posterior probabilities, jackknife support values followed by bootstrap values, respectively. Numbers between brackets on a branch that defines a clade correspond to the groups obtained with the Automatic Barcode Gap Discovery (ABGD) method for the marker.
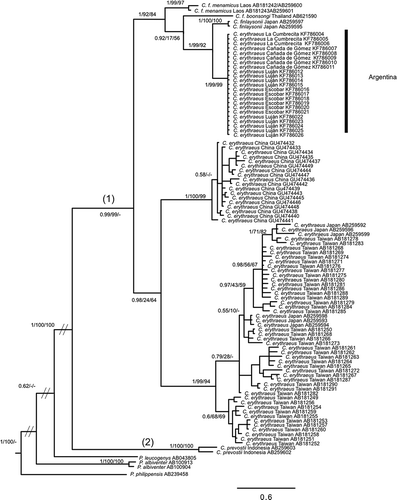
Figure 3. Bayesian inference trees of Callosciurus genus. Tree derived from cytochrome b (Cyt b). Numbers next to branches are Bayesian posterior probabilities, jackknife support values followed by bootstrap values, respectively. Numbers between brackets on a branch that defines a clade correspond to the groups obtained with the Automatic Barcode Gap Discovery (ABGD) method for the marker.
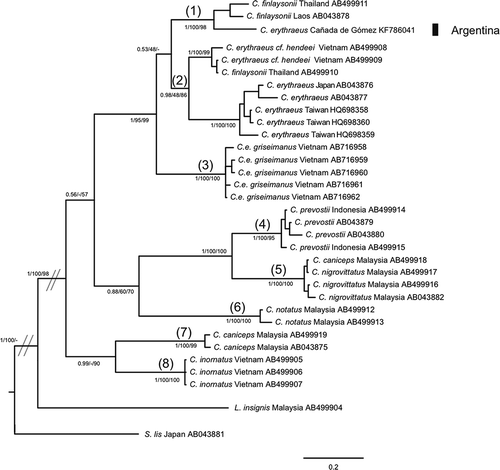
Figure 4. Bayesian inference trees of Callosciurus genus. Tree derived from citochrome oxidase c subunit I (COI). Numbers next to branches are Bayesian posterior probabilities, jackknife support values followed by bootstrap values, respectively. Numbers between brackets on a branch that defines a clade correspond to the groups obtained with the Automatic Barcode Gap Discovery (ABGD) method for the marker.
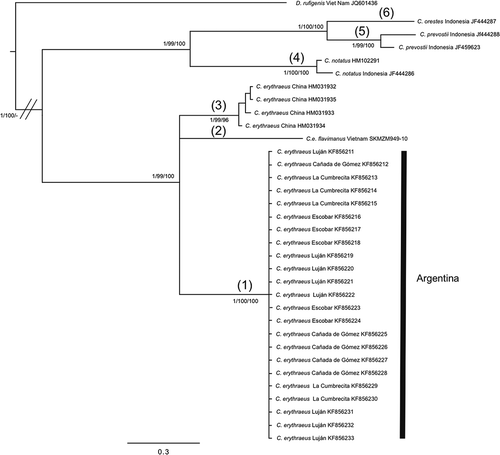
Figure 5. Bayesian inference trees of Callosciurus genus. Tree derived from recombination activating gene I (RAGI). Numbers next to branches are Bayesian posterior probabilities, jackknife support values followed by bootstrap values, respectively.
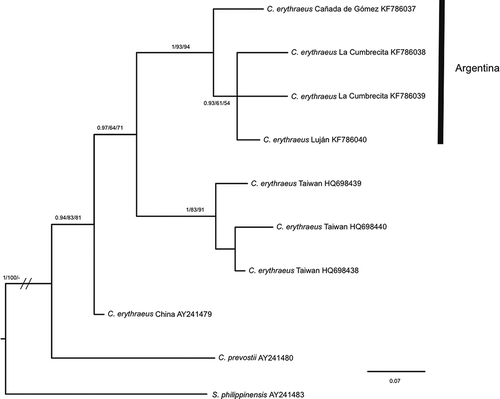
Table I. Percentage of sequence divergence (Kimura Citation1980) of Callosciurus species for the D-loop marker.
Table II. Percentage of sequence divergence (Kimura Citation1980) of Callosciurus species for the cytochrome b (Cyt b) marker.
Table III. Percentage of sequence divergence (Kimura Citation1980) of Callosciurus species for the cytochrome oxidase c subunit I (COI) marker.
Table IV. Percentage of sequence divergence (Kimura Citation1980) of Callosciurus species for the recombination activating gene I (RAGI) marker.
Polymerase chain reaction thermal cycling amplification profiles.a
Species name; locality of collection; GenBank accession numbers of markers; pelage colouration: of the belly (red, orange or creamy), other underparts such as chest, groins or genital area (red, orange or creamy), and the back (black stripe, faint black stripe or no stripe) of the squirrels captured in Argentina, and references of samples used in this study.
