Figures & data
Table 1. Antifungal activities of antagonistic bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, and negative control strain, Sphingomonas aquatilis KU408, on mycelial growth of Aspergillus candidus AC317, Aspergillus fumigatus AF8, Penicillium fellutanum KU53, and Penicillium islandicum KU101 in dual-culture assays and the effects of cell-free culture filtrates of these strains on conidial germination, germ-tube lengths, and mycelial dry weights of the fungi in potato-dextrose broth tests.
Figure 1. Antifungal activities and populations of antagonistic bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, and negative control strain Sphingomonas aquatilis KU408 against (A) Aspergillus candidus AC317, (B) Aspergillus fumigatus AF8, (C) Penicillium fellutanum KU53, and (D) Penicillium islandicum KU101, 7 d after treatment in unhulled rice grains. Sterile distilled water (SDW) and 10 mM MgSO4 solution were used as negative controls whereas a commercial fungicide, Benlate® was used as a positive control. Fungal populations were determined after 4 d of incubation at 28 °C on DG18 and total bacterial populations were evaluated after 2 d of incubation at 28 °C on nutrient agar. Different letters on the vertical bars with error bars (standard deviations, n = 6) indicate significant differences between treatments according to the least significant difference test at p < .05. CFU: colony-forming unit.
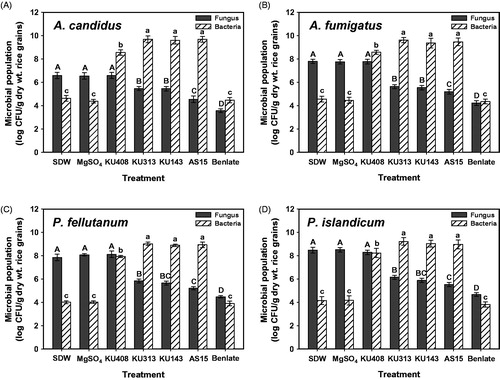
Figure 2. Photographs of antifungal activities of the volatiles produced by antagonistic bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, and negative control strain Sphingomonas aquatilis KU408 against (A) Aspergillus candidus AC317, (B) Aspergillus fumigatus AF8, (C) Penicillium fellutanum KU53, and (D) Penicillium islandicum KU101 on the I-plates. Bacterial strains or 10 mM MgSO4 solution (untreated control) were smeared on one side (nutrient agar) of the I-plates and the other side (potato dextrose agar) was inoculated with a conidial suspension of the tested fungi 24 h after the bacterial treatment. Insets are photographs of the conidial germination of the fungi affected by the bacterial volatiles or MgSO4 solution. C: conidium; gt: germ tube. Scale bar, 20 µm.
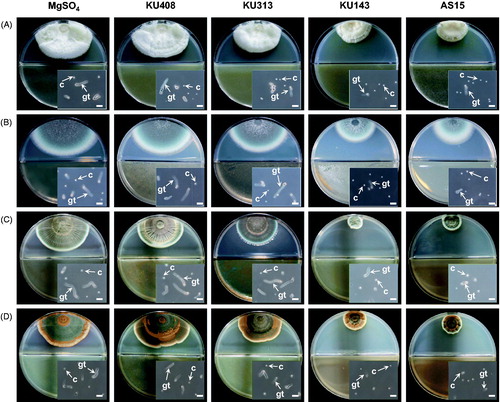
Table 2. Antifungal activities of the volatiles produced by antagonistic bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, and negative control strain, Sphingomonas aquatilis KU408, on mycelial growth, sporulation, and conidial germination of Aspergillus candidus AC317, Aspergillus fumigatus AF8, Penicillium fellutanum KU53, and Penicillium islandicum KU101 on I-plates.
Figure 3. Antifungal activities of the volatiles produced by antagonistic bacterial strains, Microbacterium testaceum KU313, Bacillus megaterium KU143, and Pseudomonas protegens AS15, and negative control strain Sphingomonas aquatilis KU408 against populations of (A) Aspergillus candidus AC317, (B) Aspergillus fumigatus AF8, (C) Penicillium fellutanum KU53, and (D) Penicillium islandicum KU101, 7 d after treatment in unhulled rice grains on the I-plates. Bacterial strains or 10 mM MgSO4 solution (untreated control) were smeared on one side (nutrient agar) of the I-plates and the other side (rice grains) was inoculated with a conidial suspension of the tested fungi, 24 h after bacterial treatment. Fungal populations in rice grains were assessed on DG18 after 4 d of incubation at 28 °C. Different capital letters on the vertical bars indicate significant (p < .05) differences between treatments according to the least significant difference test. CFU: colony-forming unit.
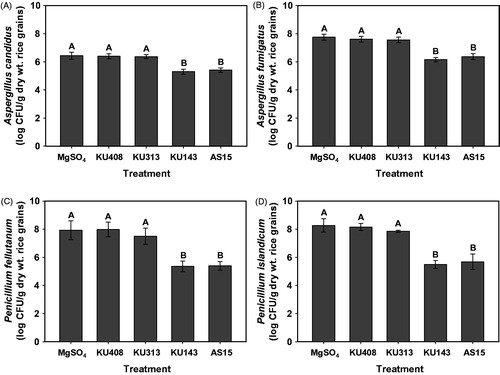
Figure 4. Gas chromatographs of the volatiles produced by (A) Bacillus megaterium KU143 and (B) Pseudomonas protegens AS15. Antagonistic bacterial strains were cultured in tryptic soy broth (TSB) at 28 °C for 24 h. Volatile compounds from B. megaterium KU143 were identified as: heptane [peak number (PN) 1], 2,5-dimethyltetrahydrofuran (PN 2), methylcyclohexane (PN 3), 3-hexanone (PN 4), 2-hexanone (PN 5), trimethyl [4-(1,1,3,3,-tetramethylbutyl) phenoxy] silane (PN 6), 3-hexene-2,5-diol (PN 7), 2-bromohexane (PN 8), 2,4,4-trimethyl-1-pentanol (PN 9), 2,6-dimethylpiperazine (PN 10), 2,2-dimethyl-3-hexanol (PN 11), and 5-methyl-2-phenyl-1 H-indole (PN 12). Volatile compounds from P. protegens AS15 were identified as: heptane (PN 1), dimethyl disulfide (PN 2), 2,2,3-trimethylpentane (PN 3), 4-methylheptane (PN 4), 3-hexanone (PN 5), 2-hexanone (PN 6), 2,4-dimethyl-1-heptene (PN 7), 3-methylcyclopentanol (PN 8), 3,3-dimethyloctane (PN 9), 2,6-dimethylnonane (PN 10), 1-(2-methylbutyl)-1-(1-methylpropyl)-cyclopropane (PN 11), 2-butyl-1-octanol (PN 12), undecane (PN 13), 1,3-di-tert-butylbenzene (PN 14), 4-trifluoroacetoxyhexadecane (PN 15), 2-isopropyl-5-methyl-1-heptanol (PN 16), and isotridecanol (PN 17). However, no distinct volatile compounds were detected in the TSB control.
![Figure 4. Gas chromatographs of the volatiles produced by (A) Bacillus megaterium KU143 and (B) Pseudomonas protegens AS15. Antagonistic bacterial strains were cultured in tryptic soy broth (TSB) at 28 °C for 24 h. Volatile compounds from B. megaterium KU143 were identified as: heptane [peak number (PN) 1], 2,5-dimethyltetrahydrofuran (PN 2), methylcyclohexane (PN 3), 3-hexanone (PN 4), 2-hexanone (PN 5), trimethyl [4-(1,1,3,3,-tetramethylbutyl) phenoxy] silane (PN 6), 3-hexene-2,5-diol (PN 7), 2-bromohexane (PN 8), 2,4,4-trimethyl-1-pentanol (PN 9), 2,6-dimethylpiperazine (PN 10), 2,2-dimethyl-3-hexanol (PN 11), and 5-methyl-2-phenyl-1 H-indole (PN 12). Volatile compounds from P. protegens AS15 were identified as: heptane (PN 1), dimethyl disulfide (PN 2), 2,2,3-trimethylpentane (PN 3), 4-methylheptane (PN 4), 3-hexanone (PN 5), 2-hexanone (PN 6), 2,4-dimethyl-1-heptene (PN 7), 3-methylcyclopentanol (PN 8), 3,3-dimethyloctane (PN 9), 2,6-dimethylnonane (PN 10), 1-(2-methylbutyl)-1-(1-methylpropyl)-cyclopropane (PN 11), 2-butyl-1-octanol (PN 12), undecane (PN 13), 1,3-di-tert-butylbenzene (PN 14), 4-trifluoroacetoxyhexadecane (PN 15), 2-isopropyl-5-methyl-1-heptanol (PN 16), and isotridecanol (PN 17). However, no distinct volatile compounds were detected in the TSB control.](/cms/asset/79617019-24c6-4471-b627-3a71584de10d/tmyb_a_1454015_f0004_c.jpg)
