Figures & data
Table 1. Cryptococcus neoformans strains used in this study.
Table 2. Oligonucleotides used in this study.
Figure 1. Phylogenetic tree depicting Mon1 in various fungal species. Sequences from Neurospora crassa OR74A (XP_959164.3), Magnaporthe oryzae 70-15 (XP_003710676.1), Trichoderma reesei QM6a (XP_006963764.1), Fusarium oxysporum Fo47 (EWZ45038.1), Gibberella zeae PH-1 (XP_011326784.1), Botrytis cinerea T4 (CCD56988.1), Aspergillus fumigatus AF293 (Q4WHL1.2), Penicillium chrysogenum (KZN85230.1), Coccidioides immitis RS (XP_001247287.2), Histoplasma capsulatum H88 (EGC46448.1), Candida albicans SC5314 (KHC78337.1), Saccharomyces cerevisiae S288c (NP_011391), Cryptococcus neoformans H99 (XP_012049217.1), Cryptococcus gattii Ru294 (KIR57394.1), Ustilago hordei (CCF52623.1), and Ustilago maydis 521 (XP_011389404.1) were compared and a phylogenetic tree of the Mon1 orthologs was generated via the MEGA 5 software (http://www.megasoftware.net/) using the alignment data from ClustalW2. The tree results were submitted to iTOL (http://itol.embl.de/) to generate the figure (left). Domain architecture of the Mon1 orthologs in fungi (right).
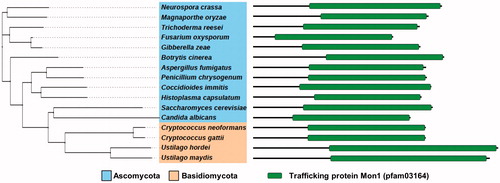
Figure 2. Phenotypes of the Cnmon1Δ mutant following exposure to various stresses. Spot dilution assays with WT (H99), Cncna1Δ (KK1), Cnmon1Δ (HP55 and HP56), and Cnmon1 + CnMON1 (HPC3 and HPC4). Cells were incubated overnight, diluted 10-fold, and plated on YPD agar. Plated cells were incubated for 2 days at 30, 37, 38, and 39 °C. (A) Serially diluted cells were also plated on YPD agar without or with (B) dithiothreitol (DTT), (C) sodium dodecylsulfate (SDS), (D) calcofluor white, (E) congo red, and (F) fluconazole at the indicated concentrations. Results shown are representative of two independent experiments.
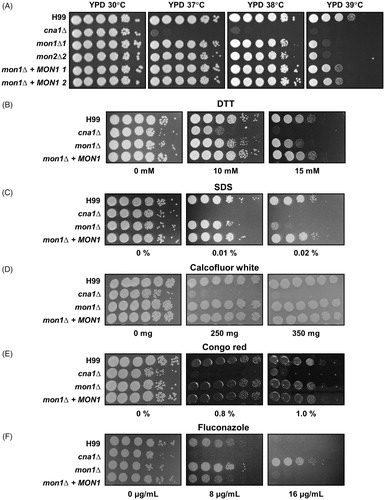
Figure 3. Virulence of the Cnmon1Δ mutant in the insect larvae model. Each Galleria larva (12 per group) was infected with approximately 80,000 WT (H99), Cncna1Δ (KK1), or Cnmon1Δ (HP55 and HP56) cells. The infected larvae were maintained at 37 °C and monitored daily for 12 days.
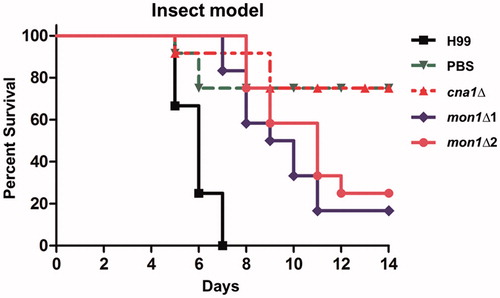
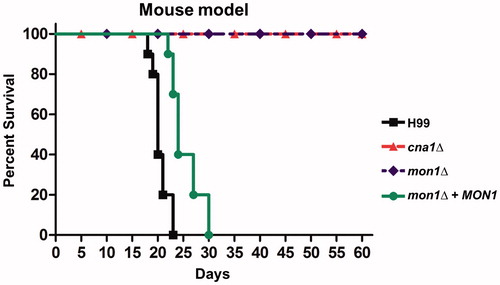
Figure 4. Virulence of the Cnmon1Δ mutant in the murine inhalation model. Cnmon1Δ mutant exhibits avirulence when compared to the WT and Cnmon1 + CnMON1 complement strains. WT (H99), Cncna1Δ (KK1), Cnmon1Δ (HP55), and Cnmon1 + CnMON1 (HPC3) cells were grown overnight in YPD broth at 30 °C and washed with PBS. Cells (5 × 105) were inoculated into A/J mice via intranasal instillation. Animal survival was monitored for 60 days post-infection.