Figures & data
Table 1. Primer sequences.
Figure 1. Seed germination (%) of tomato plants in water agar grown with the tested fungi. A total of 49 fungal isolates were inoculated on water agar amended with 0.2% glucose, and surface sterile tomato seeds were placed at the edge of the fungal mycelia. The germinated seeds and radicles were evaluated.
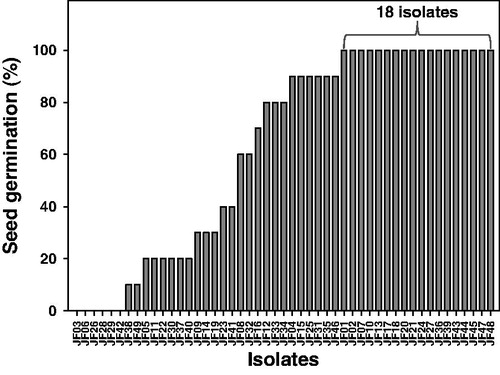
Figure 2. Growth promotion of tomato plants by four fungal isolates (JF02, JF07, JF27, and JF44). Surface sterile tomato seeds were treated with a fungal spore suspension (107 spores/mL) at 28 °C for 3 h. Tomato seeds were grown in pots containing a potting mixture for 4 weeks (5–6 leaf stage), then the shoot length (cm) and fresh weight (g) were measured. Asterisks on the bars indicate significant differences (p < .05) compared with the control according to the least significant difference test.
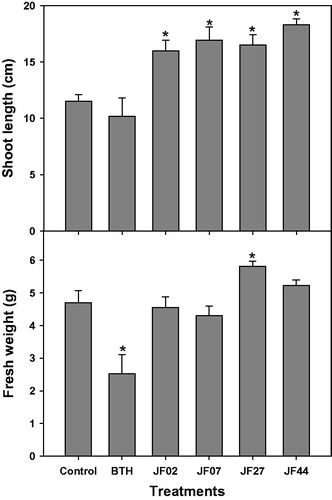
Figure 3. Resistance of tomato plants to Pseudomonas syringae pv. tomato DC 3000 induced by the fungal isolates. Seeds were treated with a fungal spore suspension, and 4 weeks after seeding, the bacterial pathogen (104 cells/mL) was infiltrated into the third leaf of each tomato plant. The infiltrated leaf was macerated and plated onto LB amended with rifampicin (50 μg/mL). Colony-forming units (CFUs) were counted. Asterisks on the bars indicate significant differences (p < .05) compared with the control according to the least significant difference test.
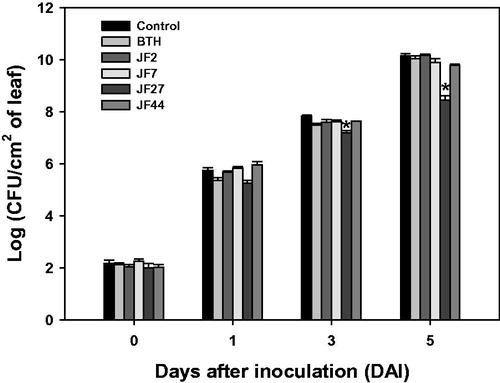
Figure 4. Relative gene expression in tomato plants after infection with Pseudomonas syringae pv. tomato DC3000. Expression analysis of genes PR1, PAL1, EIN2, and JAR1 at 0, 6, and 12 h after inoculation. Actin was used as an internal control. Asterisks on the bars indicate significant differences (p < .05) compared with the control according to the least significant difference test.
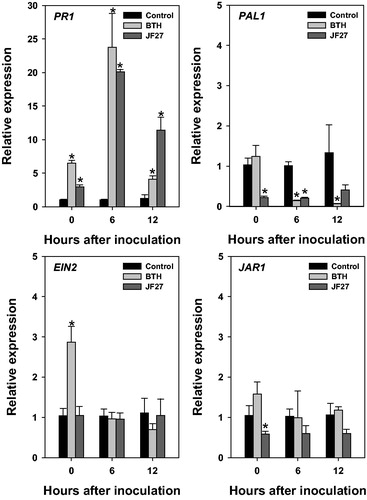
Figure 5. Phylogenetic trees created using the neighbor-joining method showing the relationships between isolate JF27 and the other members of the genus Aspergillus, based on a phylogenetic analysis of the partial nuclear ribosomal internal transcribed spacer (ITS) region and the β-tubulin sequence. Bootstrap values of 1000 analyses are shown at the branching points. The scale bar represents numbers of nucleotide substitutions per 100 nucleotides of the sequence. The type isolate is indicated as “T”; accession numbers deposited at GenBank are shown in parentheses.
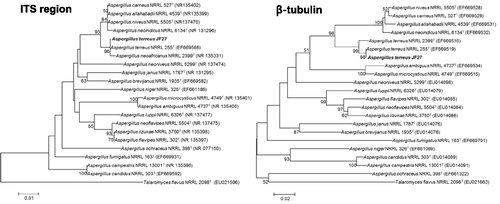
Table 2. Characterization of fungi isolates JF02, JF07, JF27, and JF44.
