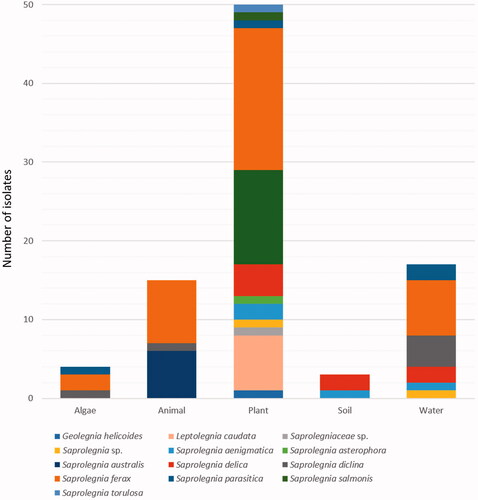
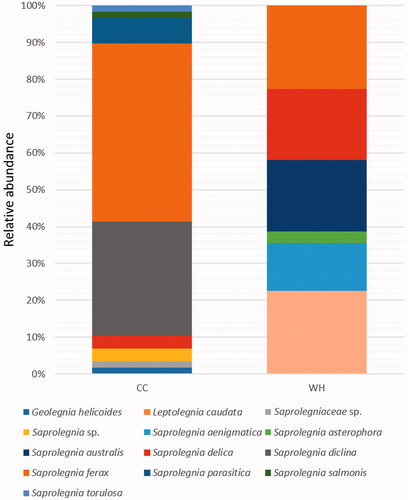
Figures & data
Figure 1. Location of the 34 sampling sites (red dot) in South Korea. The map was created using QGIS v.3.24.
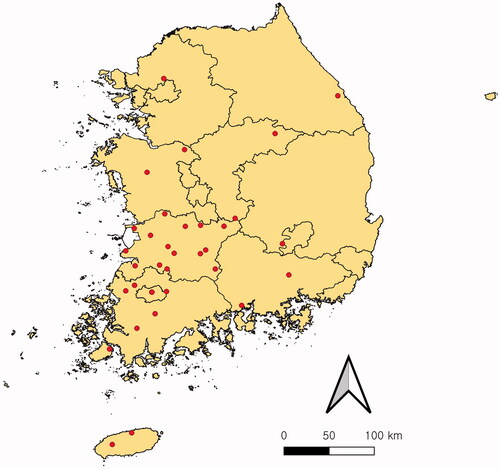
Table 1. Collection details and GenBank accession number for Saprolegniaceae isolates investigated in this study.
Figure 2. Cultural characteristics of Saprolegniaceae isolates obtained from this study. Geolegnia helicoides W732 (A); Leptolegnia caudata W1297 (B); Saprolegnia aenigmatica W1247 (C); Saprolegnia diclina W724 (D); Saprolegnia ferax W956 (E) on PDA (1, 2), V8A (3, 4), CMA (5, 6), after 72 h at 25 °C (1, 3, 5: observed view and 2, 4, 6: reverse view).
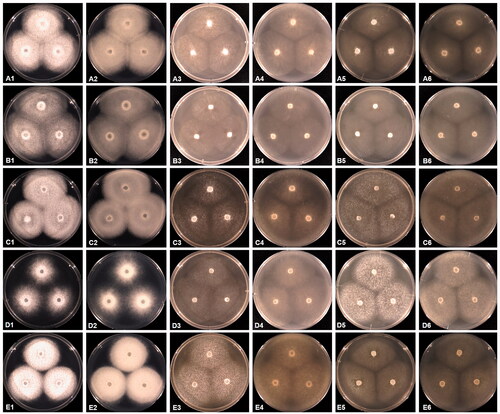
Figure 3. Morphological characteristics of Saprolegniaceae isolates obtained from the present study. Microscopic structures observed under a microscope: filiform sporangium (A) and oospore contenting finely granular (K) of Geolegnia helicoides W732, filamentous sporangium (B) of Leptolegnia caudata W1297, clavate sporangium of Saprolegnia aenigmatica W1247 (C), and S. ferax W956 (E) cylindrical sporangium (D), oogonia (G, H) and oogonia with centric oospores (L) of S. diclina W724, pyriform gemmae (F) and oogonia (J) of S. ferax W956, pyriform oogonia of S. aenigmatica W1247 (I) (scale bars: A–D, F, G, I, J, L = 20 μm, E, H, K = 10 μm).
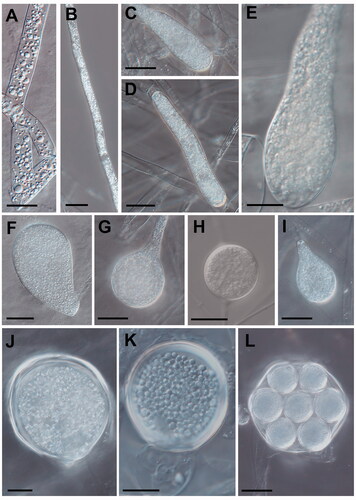
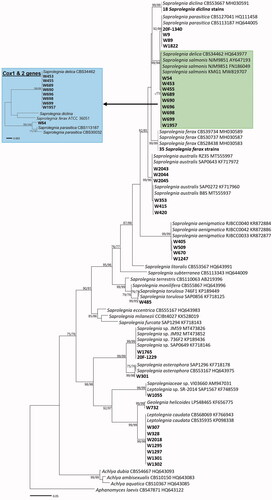
Figure 4. Phylogenetic tree of Saprolegniaceae species from the minimum evolution analysis based on the internal transcribed spacer (ITS) rDNA sequences. The large branches of Saprolegnia ferax and S. diclina were compressed. Multi-gene phylogenetic tree in a blue box was inferred using the partial cytochrome c oxidase subunit I (cox1), and cytochrome c oxidase subunit II (cox2) mtDNA sequences of taxa in a green box of the ITS tree. Bootstrapping values (minimum evolution BS/maximum-likelihood BS) higher than 70% were given above or below the branches (1000 replicates). Aphanomyces laevis was used as outgroup. The strains isolated in Korea are shown in bold. The scale bar equals the number of nucleotide substitutions per site.