Figures & data
Table 1. List of the yeast strains of Vishniacozyma terrae sp. nov. examined in the present study and related species.
Figure 1. Phylogenetic tree based on the concatenated sequences of the D1/D2 region of the LSU rRNA gene and ITS regions and constructed by the neighbor-joining method, shows relationships between strains of a novel species (YP344, YP155, and YP333) and closely related species. The novel species described in this manuscript are highlighted in bold. Saitozyma podzolica CBS 6819T was used as outgroup. Bootstrap values >50% (% of 1000 replications) were shown at branch points. Accession numbers were shown in parentheses. Bar, 0.01 substitutions per nucleotide position.
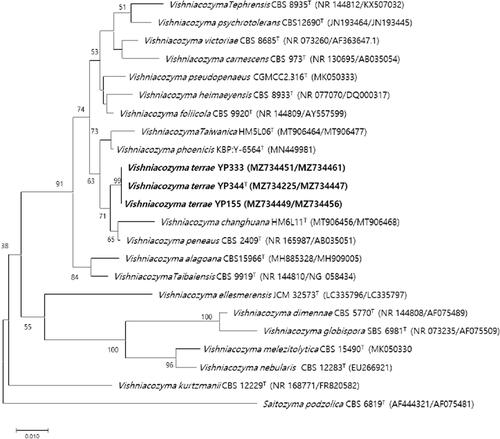
Table 2. List of the yeast strains of Dioszegia terrae sp. nov. examined in the present study and related species.
Table 3. Nucleotide substitutions in the sequences of the D1/D2 domain of the LSU rRNA gene and ITS region of Vishniacozyma terrae sp. nov. (YP344T) and Vishiniacozyma species.
Table 4. Nucleotide substitutions in the sequences of the D1/D2 domain of the LSU rRNA gene and ITS region of Dioszegia terrae sp. nov. (YP579T) and Dioszegia species.
Figure 2. Phylogenetic tree based on the concatenated sequences of the D1/D2 region of the LSU rRNA gene and ITS regions and constructed by the maximum-likelihood method, shows relationships between strains of a novel species (YP579T) and closely related species. The novel species described in this manuscript are highlighted in bold. Nielozyma formosana FK-156T was used as outgroup. Bootstrap values >50% (% of 1000 replications) were shown at branch points. Accession numbers were shown in parentheses. Bar, 0.01 substitutions per nucleotide position.
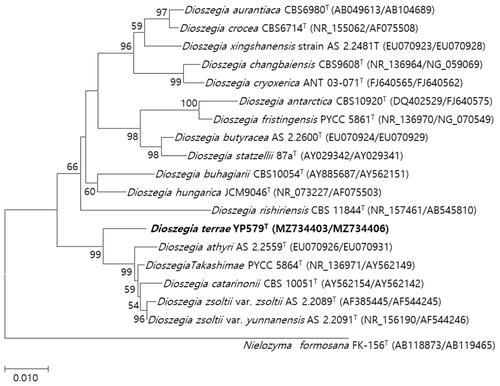
Table 5. Phenotypic characteristics that differentiate Vishniacozyma terrae sp. nov. and related species, V. peneaus and V. phoenicis.
Table 6. Phenotypic characteristics that differentiate Dioszegia terrae sp. nov. and related species, D. zsoltii and D. catarinonii.