Figures & data
Table 1. GenBank accession numbers of fungal strains used in this study for phylogenetic analysis.
Figure 1. Cultural and morphological characteristics of Tolypocladium globosum (KNUF-22-14AT). Colonies on potato dextrose agar (A); malt extract agar (B); and cornmeal agar (C) following 14 days of incubation at 25 °C, respectively. Phialides (D–H); conidia (I,J). Scale bars: D = 20 µm, E–J = 10 µm.
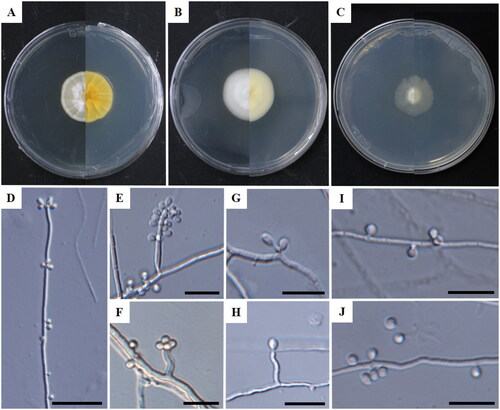
Table 2. Morphological comparison of Tolypocladium globosum (KNUF-22-14AT) with the closest species of Tolypocladium.
Figure 2. Neighbor-joining phylogenetic tree based on the concatenated sequences of ITS sequences showing the phylogenetic position of strains KNUF-22-14AT and KNUF-22-15A among Tolypocladium species. Bootstrap values based on 1000 replicates are shown next to the branches. Aschersonia confluens BCC 7961 comprised the outgroup. Bootstrap values greater than 50% (based on 1000 replications) are shown at the branching points. The isolated strains are shown in bold. Bar = 0.01 substitutions per nucleotide position.
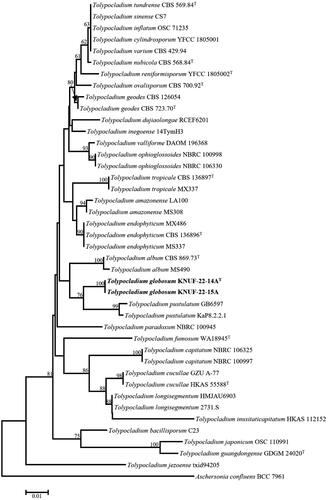
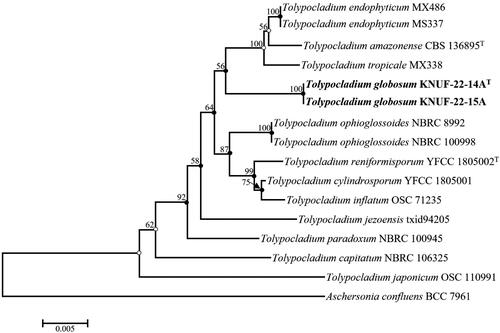
Figure 3. Neighbor-joining phylogenetic tree based on the combined molecular markers of the internal transcribed spacer (ITS) regions, small subunit (SSU), and large subunit (LSU) showing the phylogenetic position of strains KNUF-22-14AT and KNUF-22-15A among Tolypocladium species. Aschersonia confluens BCC 7961 comprised the outgroup. Bootstrap values greater than 50% (based on 1000 replications) are shown at the branching points. The neighbor-joining tree, maximum likelihood, and maximum parsimony trees indicated with filled nodes and open circles were made using maximum likelihood or parsimony. The isolated strains are shown in bold. Bar = 0.005 substitutions per nucleotide position.