Figures & data
Table 1. Aspergillus genome sequences used in this study.
Table 2. Degrees of sequence identity of PWE36 aflatoxin biosynthesis genes to those of Aspergillus sojae strains and Aspergillus parasiticus isolates.
Table 3. Degrees of sequence identity of PWE36 developmental genes to those of Aspergillus sojae strains and Aspergillus parasiticus isolates.
Figure 1. Alignment of portions of pksA/aflC sequences (A); aflR sequences (B) from Aspergillus parasiticus isolates, PWE36, and Aspergillus sojae strains. SNPs in blue that yield pre-termination codons in pksA/aflC (TAA) and aflR (TGA) genes, respectively, are yellow highlighted. Numbers correspond to nucleotide positions in A. sojae gene sequences.
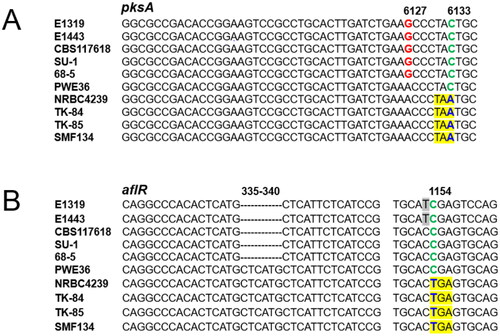
Figure 2. Schematic representations of deletions in the CPA gene clusters of Aspergillus parasiticus isolates, PWE36, and Aspergillus sojae strains. The CPA gene cluster of Aspergillus flavus AF36 (16.8 kb) was used as the alignment template. The comparisons on the lower panel were drawn to scale. Blank space indicates deleted portions. Grey lines are nonhomologous regions of replacement and the 112 bp deletions. Thick red lines are highly homologous sequences.
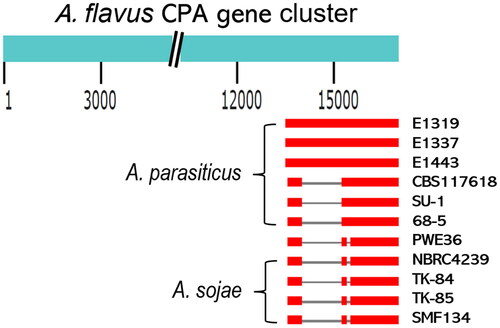
Figure 3. Schematic representation of homologous regions in genome sequences of Aspergillus sojae strains, PWE36, and Aspergillus parasiticus isolates. The genome sequence of A. sojae SMF134 that had been assembled at the chromosome level was used as the reference; it was concatenated in chromosome order (from I to VIII). Colored blocks are locally collinear blocks (LCBs), which indicate homologous regions between two genomes. The LCB weight was set at around 550 except for the SMF134/TK-84 pair, of which the lowest score was 24,283. Regions that were split and translocated in a compared genome are shown as inverted segments at the bottom.
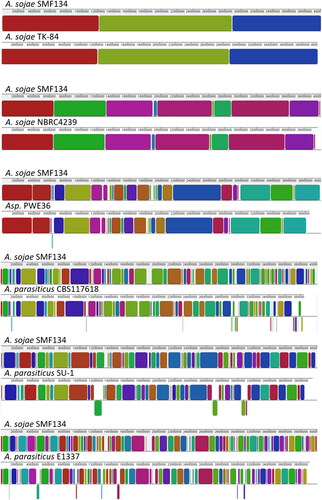
Figure 4. (A) Total SNP counts from paired genome sequence comparisons among PWE36, Aspergillus sojae strains, and Aspergillus parasiticus isolates; (B) Phylogenetic tree inferred from concatenated sequences of total SNPs by the Neighbor-Joining method with 100 bootstrap iterations. The number of total SNP count for each sequence was 618,048. The estimated divergence time (1.1 mya) between A. parasiticus SU-1 and A. sojae NRBC4239 was derived from the reference divergence time scale (3.8 mya) between Aspergillus flavus NRRL3357 and Aspergillus oryzae RIB40 [Citation38].
![Figure 4. (A) Total SNP counts from paired genome sequence comparisons among PWE36, Aspergillus sojae strains, and Aspergillus parasiticus isolates; (B) Phylogenetic tree inferred from concatenated sequences of total SNPs by the Neighbor-Joining method with 100 bootstrap iterations. The number of total SNP count for each sequence was 618,048. The estimated divergence time (1.1 mya) between A. parasiticus SU-1 and A. sojae NRBC4239 was derived from the reference divergence time scale (3.8 mya) between Aspergillus flavus NRRL3357 and Aspergillus oryzae RIB40 [Citation38].](/cms/asset/9611a16f-81bc-4e09-a194-705029b4f653/tmyb_a_2217495_f0004_c.jpg)
