Figures & data
Table 1. List of fungal isolates obtained from soybean seeds with decay symptom.
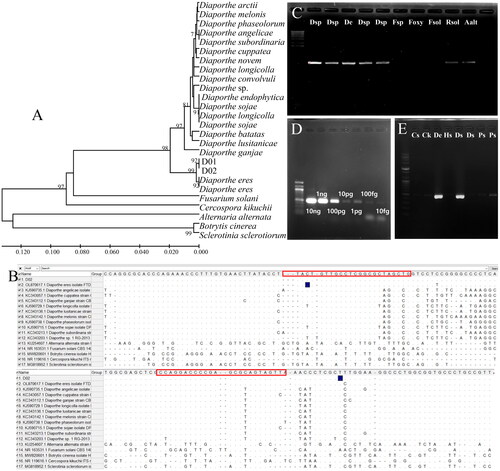
Figure 1. Maximum likelihood phylogenetic analysis of the Diaporthe species using ITS primers to develop a specific marker to detect Diaporthe spp. The bootstrap numbers represent the percent of 1000 replicates (only values greater or equal to 70% are shown) (A); specific detection of Diaprothe spp. from seven isolates, including Cercospora kikuchii, B. cienrea, F. solani, Sclerotinia sclerotiorum, and Alternaria alternata (B); lane M, 1000 bp DNA ladder; lanes dsp, Diaporthe sp.; lane De, D. eres; lane fsp, Fusarium sp.; lane foxy, F. oxysporum; lane fsol, F. solani; lane, R. solani; and lane aalt, A. alternata (C); Conventional PCR amplification of different amounts of Diaporthe eres DNA with the primers D_sp3F3R. M, 100 bp ladder marker; 10 ng; 1 ng; 100 pg; 10 pg; 1 pg; 100 fg; 10 fg. (D); detection of Diaporthe spp. using D_sp3F3R from Cercospora sojina, C. kikuchii, D. eres, and soybean seeds with decay and discoloration; lane Cs, C. sojina C01; lane Ck, C. kikuchii; lane De, D. eres D01; lane Hs, healthy seed; lanes Ds, seeds with decay symptoms; lanes Ps, brown and purple seeds. (E).
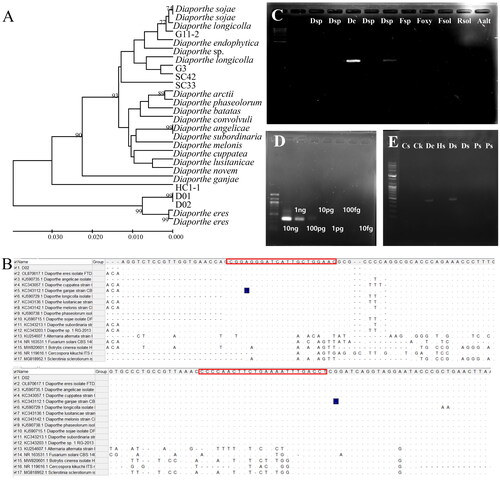
Table 2. Information on primers used in this study.
Figure 2. Maximum likelihood phylogenetic analysis of the Diaporthe species using ITS primers to develop a specific marker to detect D. eres. The bootstrap numbers represent the percent of 1000 replicates (only values greater or equal to 70% are shown) (a); specific detection of D. eres from seven isolates, including Cercospora kikuchii, B. cienrea, F. solani, Sclerotinia sclerotiorum, and Alternaria alternata (B); lane M, 1000 bp DNA ladder; lanes 1 and 2, Diaporthe sp.; lane 3, D. eres; lanes 4 and 5, Diaporthe sp.; lane 6, Fusarium sp.; lane 7, F. oxysporum; lane 8, F. solani; lane 9, R. solani; and lane 10, A. alternata (C); Conventional PCR amplification of different amounts of D. eres DNA with the primers D_eres 3F3R. M, 100 bp ladder marker; 1, 10 ng; 2, 1 ng; 3, 100 pg; 4, 10 pg; 5, 1 pg; 6, 100 fg; 7, 10 fg. (D); detection of Diaporthe spp., D_sp3F3R from Cercospora sojina, C. kikuchii, D. eres, and soybean seeds with decay and discoloration; lane a, C. sojina C01; lane b, C. kikuchii; lane c, D. eres D01; lane d, healthy seed; lanes e and f, seeds with decay symptoms; lanes g and h, brown and purple seeds. (E).