Figures & data
Table 1. List of powdery mildew samples collected in Korea.
Figure 1. Primer design on GAPDH sequences of Golovinomyces species. The newly designed primers (GoGPD-F and GoGPD-R) are shown above and below blue bars, while the previously used primers (PMGAPDH1 and PMGAPDH3R) are shown above and below red bars.

Table 2. Sequence analysis of Golovinomyces species using the newly developed GAPDH primers and the ribosomal ITS and LSU primers.
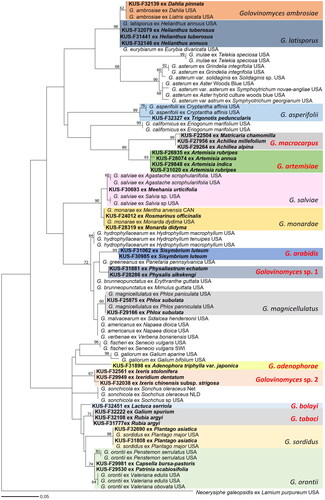
Figure 2. Maximum likelihood tree of Golovinomyces species based on GAPDH sequences. Bootstrapping support values higher than 60% are given at the branches. Korean specimens sequenced in this study are in bold. Golovinomyces species initially sequenced for the GAPDH gene in this study are highlighted in red. The scale bar equals the number of nucleotide substitution sites.