Figures & data
Figure 1. Standardized principal component analysis of the variation of three functional traits in seven weed species, measured at the individual level within three field elements. (A) Contribution of traits to the first and second principal components. (B) Location of plants sampled from different field elements on the ordination plan; 90% confidence ellipses around field element centroids are drawn. (C) Location of plants belonging to different species on the ordination plan. EPPO codes refer to abbreviations as presented in Table .
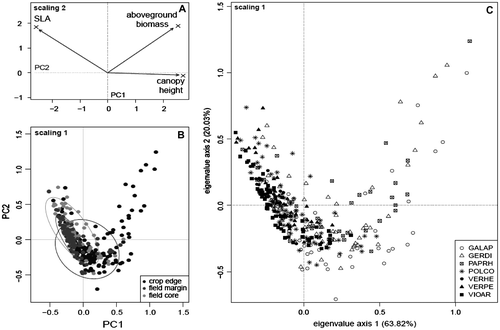
Table 1. Linear mixed model analyses on functional trait variation for seven weed species in three field elements (crop edge, field margin and field core). Tukey’s post hoc tests investigate the pairwise differences between field elements. Square-root (SLA and canopy height) or log10-transformations (above-ground biomass) were applied to meet the assumptions of homogeneity of variance (Bartlett test) and normality of the residuals (Shapiro–Wilk test). *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant.
Figure 2. Distribution of trait values per species and field element. The horizontal line on each beanplot is the mean value of the distribution. The graphical representation is based on a non-parametric Gaussian kernel density estimation. The canopy height and the specific leaf area were square-root transformed while the above-ground biomass was log10-transformed. CE, crop edge; FM, field margin; FC, field core. EPPO codes refer to abbreviations as presented in Table
Table 2. Interspecific comparisons of trait distribution within each field element. Overlap in functional trait values between weed species sampled within (A) the crop edge and (B) the field margin. Non-parametric Gaussian kernel density estimation was used to measure functional overlap for each trait (top). 1 – Fβ is a measure of functional overlap in the multi-trait space (bottom). Values in bold are significantly lower than under the randomization null model after applying sequential Bonferroni–Holm adjustment (P < 0.05). Abbreviations used to represent species refer to EPPO codes as follow: Galium aparine (GALAP), Geranium dissectum (GERDI), Papaver rhoeas (PAPRH), Fallopia convolvulus (POLCO), Veronica hederifolia (VERHE), Veronica persica (VERPE) and Viola arvensis (VIOAR).
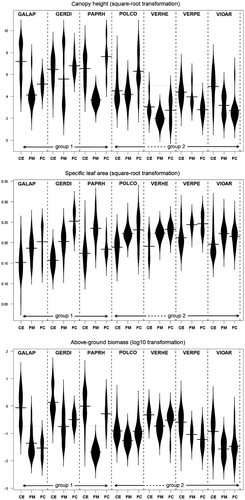
Table 2. Interspecific comparisons of trait distribution within each field element. Overlap in functional trait values between weed species sampled within (A) the crop edge and (B) the field margin. Non-parametric Gaussian kernel density estimation was used to measure functional overlap for each trait (top). 1 – Fβ is a measure of functional overlap in the multi-trait space (bottom). Values in bold are significantly lower than under the randomization null model after applying sequential Bonferroni–Holm adjustment (P < 0.05). Abbreviations used to represent species refer to EPPO codes as follow: Galium aparine (GALAP), Geranium dissectum (GERDI), Papaver rhoeas (PAPRH), Fallopia convolvulus (POLCO), Veronica hederifolia (VERHE), Veronica persica (VERPE) and Viola arvensis (VIOAR).
Table 3. Intraspecific comparisons of trait distribution between field elements. Overlap in functional trait values between plants sampled from different field elements within each weed species. Non-parametric Gaussian kernel density estimation was used to measure functional overlap for each trait (top). 1 – Fβ is a measure of functional overlap in a multi-trait space (bottom). Values in bold are significantly lower than under the randomization null model assumption after applying sequential Bonferroni–Holm adjustment (p < 0.05). CE, crop edge; FM, field margin; FC, field core; NA, data not available due to too small sample sizes. EPPO codes refer to abbreviations as presented in Table .
