Figures & data
Table 1. Synoptic table of the populations studied reporting the field identification, location, habitat characteristics and voucher details.
Figure 1. Principal components analysis based on the seven considered morphological characters, with groups corresponding to the eight populations. PC1: Eigenvalue 55.81, % variance 81.50; PC2: Eigenvalue 8.17, % variance 11.93. Yellow: Chebba; magenta: Zembra; red: Lampedusa; light green: Linosa 1; dark green: Linosa 2; brown: Cannatello; dark blue: Marsala, light blue: Balestrate.
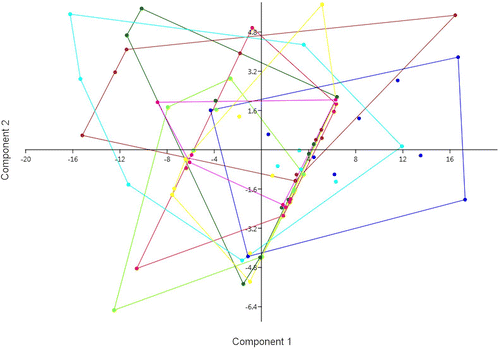
Figure 2. Discriminant analysis based on the seven considered morphological characters, with groups corresponding to the eight populations. Axis 1: Eigenvalue 0.40153, % variance 38.6; Axis 2 Eigenvalue 0.32117, % variance 30.88. Yellow: Chebba; magenta: Zembra; red: Lampedusa; light green: Linosa 1; dark green: Linosa 2; brown: Cannatello; dark blue: Marsala, light blue: Balestrate.
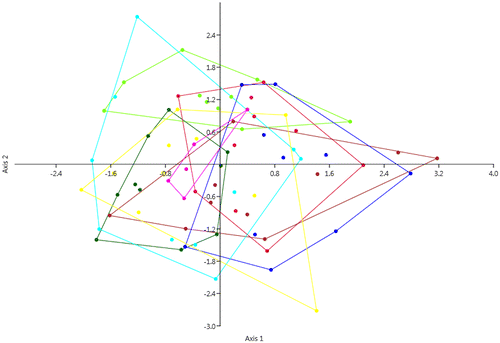
Figure 3. Plots of the six continuous numeric characters. For each sample, the 25–75% quartiles are drawn using a box. The median is shown with a horizontal line inside the box. The whiskers are drawn from the top of the box up to the largest data point less than 1.5 times the box height from the box, and similarly below the box. Outliers values are shown as stars. No significant differences among populations are highlighted. T(white): Overall; 1(yellow): Chebba; 2(magenta): Zembra; 3(red): Lampedusa; 4(light green): Linosa 1; 5(dark green): Linosa 2; 6(brown): Cannatello; 7(dark blue): Marsala, 8(light blue): Balestrate.
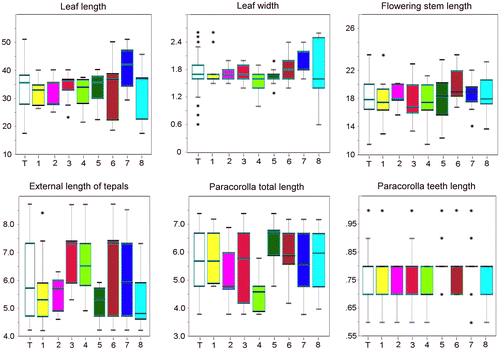
Table 2. Barcoding polymerase chain reaction primers adopted in this study.
Table 3. Reference data of the newly generated sequences and the subset of the related sequences for each taxon considered in this study.
Figure 4. Phylogenetic neighbour-joining tree generated from the most significant matK gene sequences listed in Table .
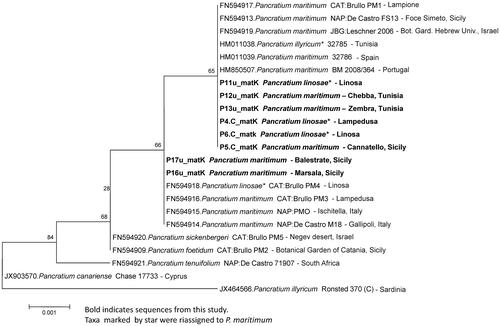
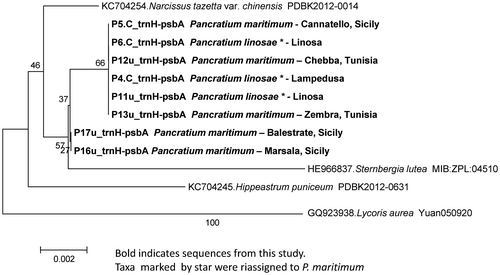
Figure 5. Phylogenetic neighbour-joining tree of the most significant trnH-psbA IGS sequences listed in Table .