Figures & data
Table 1. Sequence similarity comparison at the nucleotides and amino acids (within parentheses) sections for the signal peptide (SP), α1, α2 and connecting peptide/transmembrane/cytoplasmatic (CP/TM/CY) domains of the DRA genes from different species compared to those for gayal (B. frontalis, up) and gaytle (B. frontalis × B. taurus, down).
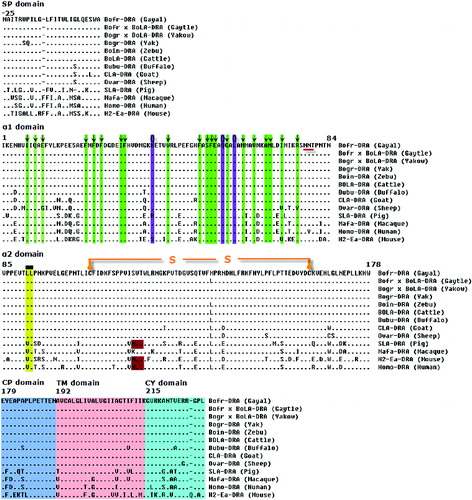
Figure 2. Alignment of the protein encoded by the DRA genes in gayal (Bos frontalis), gaytle (B. frontalis × B. taurus), European cattle (B. taurus, D37956), zebu cattle (Bos indicus, FM986338), yak (Bos grunniens, JQ911700), Chinese yakow (B. grunniens × B. taurus, JQ347519), buffaloes (Bubalus bubalis, DQ016629), goats (Capra hircus, AB008754), sheep (Ovis aries, M73983), macaques (Macaca mulatta, EF208826), humans (Homo sapiens, NM_019111), pigs (Sus scrofa, M93028) and mice (Mus musculus, NM_010381). A hyphen (-) indicates amino acid identity and an asterisk (*) indicates a gap inserted for maximum alignment. Arrows (↓) within the green column indicate the amino acid positions constituting part of peptide-binding site (PBS). A circle (О) with a blue column indicates a conserved site for T-cell receptor interaction and putative N-linked glycosylation sites are underlined. A rectangle sign (![]()