Figures & data
Figure 1. Complementary annealing mediated by exonuclease (CAME) for seamless cloning. (A) Insert DNA is amplified using PCR primers tailed by the sequences of more than 15 bp complementary to the backbone vector. Linearized vector can be generated by either restriction enzymatic digest or PCR. Enzymes either with 3′→5′ or 5′→3′ exonuclease activity are used to create single strand overhang. The two substrates are joined through annealing. (B) When PCR amplification of insert is difficult, insert can be generated by restriction enzyme digest, but the resulted fragment likely carries undesired sequences. Vector is then amplified using PCR primers containing sequences complementary to the insert. A sequential treatment with 5′→3′ exonuclease and 3′→5′ exonuclease ensures the formation of nicked circle. The gapped (A) or nicked (B) circle can be repaired after transformed into host cells.
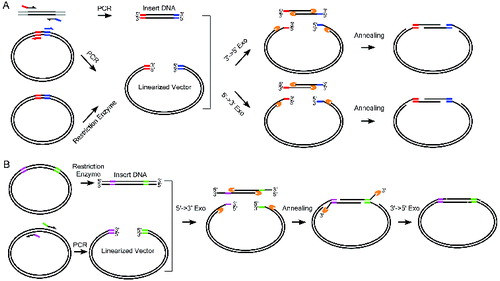
Figure 2. Test the efficiency of CAME cloning. (A) A 450 bp DNA fragment was incubated with indicated enzymes for various time periods. The weaker intensity of the bands resolved on agarose gel is correlated with the enzymatic reaction for longer period. C, the DNA without enzymatic treatment. (B) CAME cloning of a 1.1 kb insert using different enzymes. (C) Upper diagram illustrates the cloning of a 12 kb insert via CAME. Seven randomly picked white colonies (lane 1–7) were first screened by PCR amplifying the 12 kb insert and then verified by sequencing. NC, negative control using blue colony as template for PCR and PC, positive control using λ DNA as template for PCR. (D) Scheme for cloning of a 1.4 kb insert out of a DNA mixture.
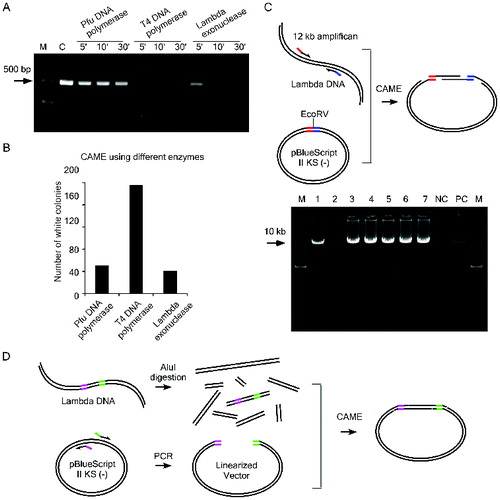
Table 1. Cloning a single DNA fragment out of a complex sample.
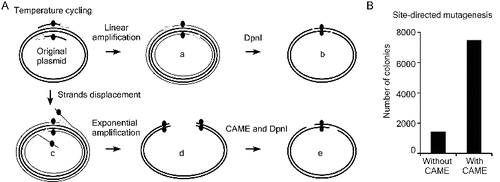
Figure 3. Improving site-directed mutagenesis using CAME. (A) Thermostable DNA polymerases possess weak strand displacement activity, which results in strand displacement (c) and exponential amplification of erroneous products (d) during site-directed mutagenesis. Such products will anneal and interfere with correct mutant DNA (b) to further reduce the mutagenesis efficiency. These erroneous products (d) can be converted into correct mutant (e) via CAME and DpnI treatment steps. (B) CAME improves site-direct mutagenesis by approximately fivefold in a point mutation of siCHECK-2.