Figures & data
Table 1. AD primers used in this study.
Table 2. Nested specific primers used in this study.
Table 3. Settings for TAIL-PCR.
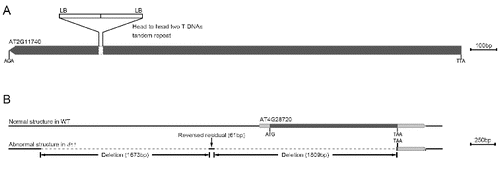
Figure 1. Four T-DNA insertion sites were cloned by TAIL-PCR in the v1 mutant. AD1-1 and AD1-2 denote two individual DNA fragments amplified by using AD1. Brackets indicate the length of the flanking sequence.

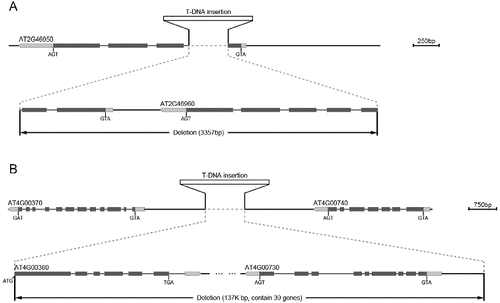
Figure 2. Flanking sequences cloned by TAIL-PCR. Arrows represent the position of the nested specific primers in the T-DNA border. Brackets show the length of flanking sequence. ×10-Lb2 represents the flanking sequences cloned by using three nested primers Lb0, Lb1 and Lb2 in the ×10 mutant. ×10-Lb3 represents the sequences cloned by using Lb1, Lb2 and Lb3 in the ×10 mutant. di2-Lb2 represents the sequences cloned by using Lb0, Lb1 and Lb2 in the di2 mutant.