Figures & data
Figure 1. Gel electrophoresis pattern of faecal and hair genomic DNA from R. brelichi. Note: Hind III digested λDNA Marker (1); negative control of extraction (2); hair DNA sample (3); faecal DNA samples (4–6).
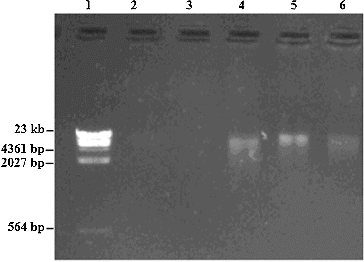
Figure 2. PCR amplification of R. brelichi mitochondrial genes. Note: PCR amplification of R. brelichi mitochondrial genes (A); DNA Marker (1); negative control of amplification (2); negative control of extraction (3); faecal DNA samples (4, 5); hair DNA sample (6); PCR amplification of R. brelichi genes (B); marker (1); negative control of amplification (2); faecal DNA samples (3–6).
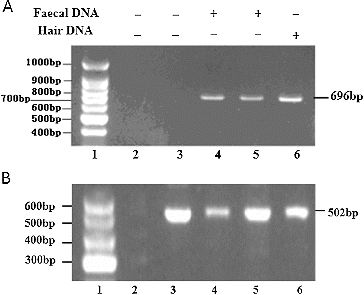
Figure 3. Partial sequencing result of R. brelichi's mitochondrial gene. Note: This is a partial sequencing result from R. brelichi mitochondrial gene, showing the sequencing quality.
Figure 4. DNA sequence alignment of mitochondrial genes (ND6, tRNA-Glu and Cyto B genes). Note: The sequence of mitochondrial genes (including ND6, tRNA-Glu and Cyto B) of R. brelichi (underlined) was aligned with those from other closely related species. Dash line indicates the same base, whereas lowercase letter shows a different base at that position. These sequences can be retrieved from NCBI.
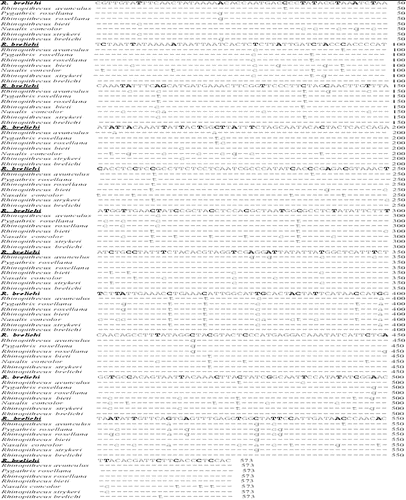
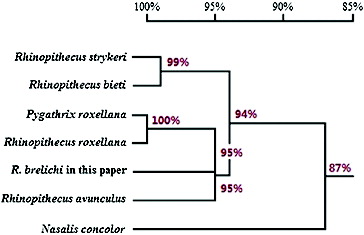
Figure 5. Homology tree from DNA sequencing of mitochondrial genes. Note: This homology tree was generated with DNAMAN12.0 program. Based on the partial sequence of the mitochondrial genes that we sequenced, we observed the closest relationship between R. brelichi, Pygathrix roxellana, Rhinopithecus roxellana and Rhinopithecus avunculus (at least 95% identity for this region). The relationship between R. brelichi and Nasalis concolor was less–only 87% of the sequences were identical.