Figures & data
Table 1. PCR primers used in this study.
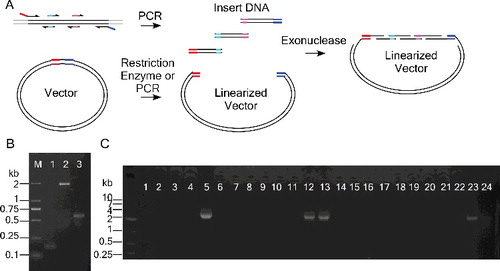
Figure 1. Use of exonuclease to improve multiple site-directed mutagenesis (MSDM). (A) Partially overlapping primers are used to amplify mutated DNA fragments with homologous ends. Exonuclease treatment helps the complementary annealing of these homologous ends. (B) Mutagenesis reaction carried out by the QuikChange method (QCMSDM) or partially overlapping primers as depicted in (A), 100 ng and 1 pg of pGL3-Olig2 plasmid were, respectively, used as a template in the two reactions. The desired bands are indicated by arrows; an additional band is indicated by an arrowhead. (C) Mutagenesis reaction was performed using 1 μL of pGL3-Olig2-containing bacterial culture. (D) T4 DNA polymerase treatment increased the yield of colonies for mutagenesis. PCR products were treated with T7 exonuclease (T7) or T4 DNA polymerase (T4) before transformation. Colony numbers are shown as means and standard deviation of three independent experiments. (E) Sequencing verification of mutagenesis.
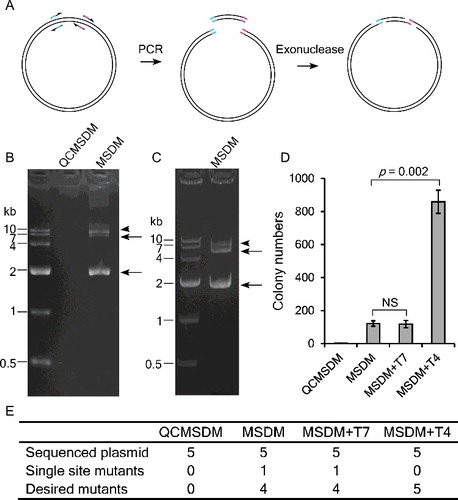
Figure 2. Products generated in MSDM PCR. During the PCR, the undesired single mutation products (b) can be used by primers and megaprimers to generate desired products (a and c), which in turn inhibits the amplification efficiency of single mutation products (b).
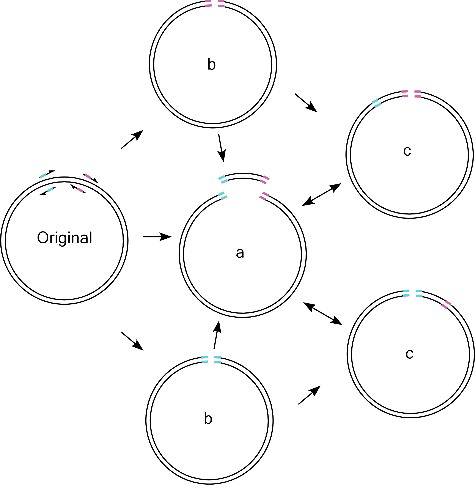
Figure 3. Cloning coupled with mutagenesis. (A) Shorter mutation-containing fragments with overlap ends are amplified in parallel. The overlapping sequences can be annealed by exonuclease treatment. (B) PCR products amplified using the three primer pairs: Lane 1, Olig2F and Olig2MR1; Lane 2, Olig2MF1 and Olig2MR2; Lane 3, Olig2MF2 and Olig2R. (C) Colony PCR using the primers of Olig2F and Olig2R. Colonies 5, 12, 13 and 23 were verified to contain the desired sequence.