Figures & data
Table 1. Oligonucleotide primers.
Figure 3. PCR and Southern blotting analysis of the ku70 gene disruption vector. PCR analysis (A) using primer pairs P14 and P15: lane 1, molecular size marker (Sangon Biotech, Shanghai, China); lane 2, wild-type strain; lane 3, ΔAcku70 strain. Southern blotting analysis (B) of the genomic DNA of the ΔAcku70 strain: lane 1, digestion using EcoRI; lane 2, digestion using SacI.
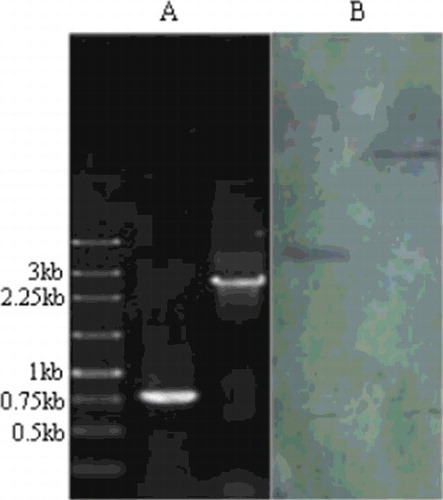
Table 2. Efficiency of gene replacement at the veA and flbA loci in wild-type and ΔAcku70 strains.