Figures & data
Table 1. Parameters and variables used in OFAT approach.
Table 2. Values of central composite design (CCD) variables.
Table 3. CCD design matrix for RSM and ANN studies in the cultivation of P. pentosaceus RF-1.
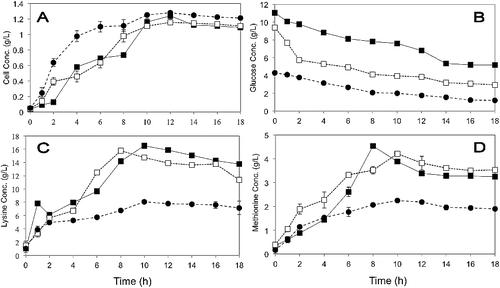
Figure 1. Optimization by OFAT on P. pentosaceus RF-1 responses to produce lysine and methionine. (A) Effect of molasses (♦) 1 g/L, (■) 3 g/L, (▴) 5 g/L, (○) 10 g/L, (◊) 12 g/L; (B) effect of nitrogen sources (♦) yeast extract, (□) peptone (▴) PKC (■) fish meal; (C) effect of fish meal (♦) 1 g/L, (■) 3 g/L, (▴) 5 g/L, (∆) 10 g/L, (□) 15 g/L, (•) 20 g/L; (D) effect of glutamic acid (♦) 0.1 g/L, (■) 0.3 g/L, (▴) 0. 5 g/L, (•) 1 g/L, (○) 5 g/L; (E) effect of initial medium pH (♦) pH 5, (■) pH 6, (▴) pH 7, (○) pH 8.
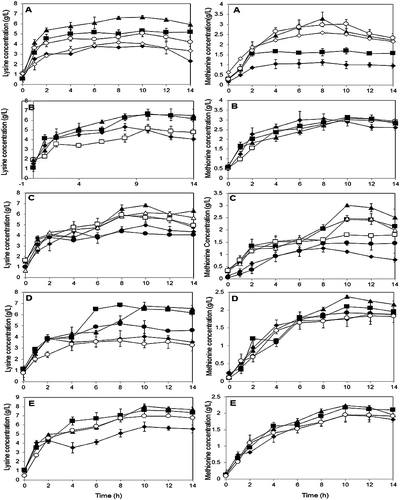
Table 4. Regression model from ANOVA on lysine–methionine biosynthesis.
Table 5. Observed, predicted, and absolute deviation of lysine–methionine biosynthesis by ANN.
Table 6. The effect of different normal feed-forward network architectures on the model residual error in the prediction of lysine–methionine biosynthesis by P. pentosaceus RF-1.
Figure 2. Finalized topology of neural network architecture (4-5-2) trained by incremental back propagation (IBP) for the estimation of lysine–methionine biosynthesis.
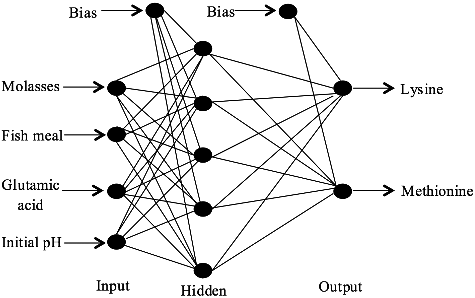
Figure 3. The level of importance of effective medium constituents on lysine–methionine biosynthesis.
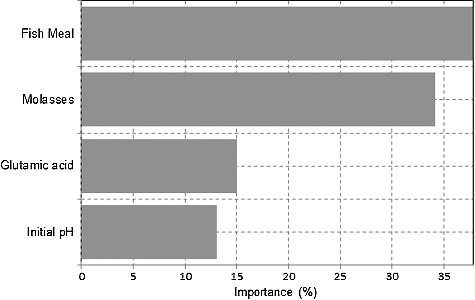
Figure 4. 3D response surface plot for lysine biosynthesis with the interaction between (a) molasses with fish meal, (b) initial pH with molasses, (c) glutamic acid with molasses, and (d) glutamic acid with fish meal as modelled by ANN.
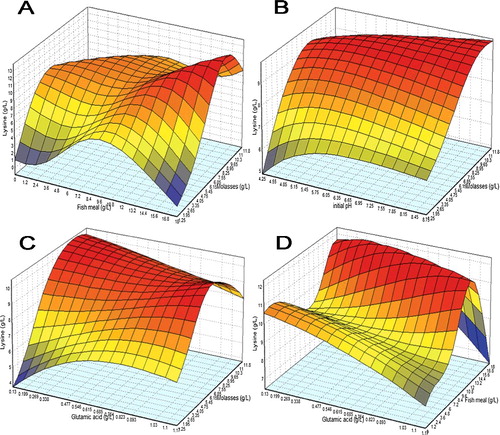
Figure 5. 3D response surface plot for methionine biosynthesis with the interaction between (a) molasses with fish meal, (b) initial pH with molasses, (c) glutamic acid with molasses, and (d) glutamic acid with fish meal as modelled by ANN.
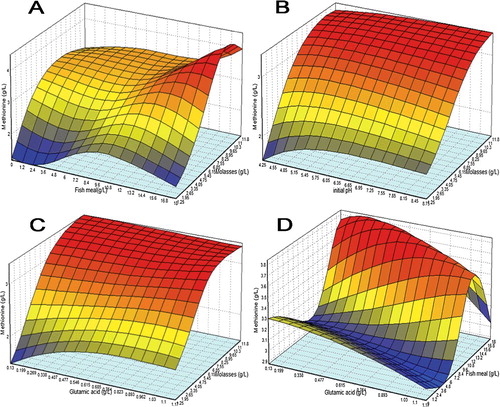
Table 7. Validation results of medium formulation and medium pH as suggested by ANN and RSM model with comparison to OFAT.