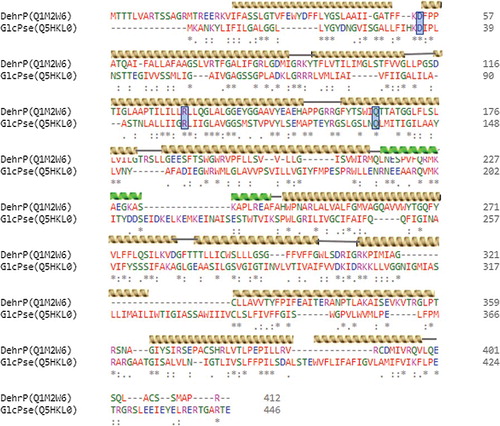
Figures & data
Figure 2. Overview of DehrP structural arrangement. (A) Side-view representation of the 12 TMs as two-fold pseudosymmetrical domains connected by a relatively large cytoplasmic α-helical loop between TM6 and TM7. (B) Periplasmic-view showing the cavity that leads into the periplasmic space with TM7 at the middle, while TMs 3, 6, 9, and 12 farther away from the cavity. (C) Cytoplasmic-face showing the cavity that leads into the cytoplasm with TM7 at the middle, while TMs 3, 6, 9, and 12 farther away from the cavity.
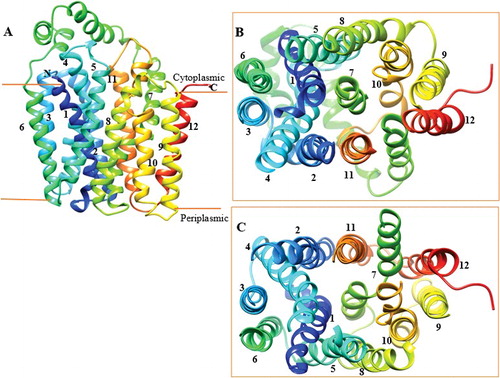
Figure 3. Structural alignment of DehrP. (A) Superposition of DehrP (blue) and GlcPse (yellow) in a schematic representation. (B) Superposition of Gln165 of DehrP on the glucose binding site residue (Gln137) of GlcPse. (C) Gln133, Asp36 and Arg130 of DehrP superposition on the proposed H+-binding site residues (Ile105, Asp22 and Arg102) of GlcPse. The grey box shows the domain region including the catalytic region.
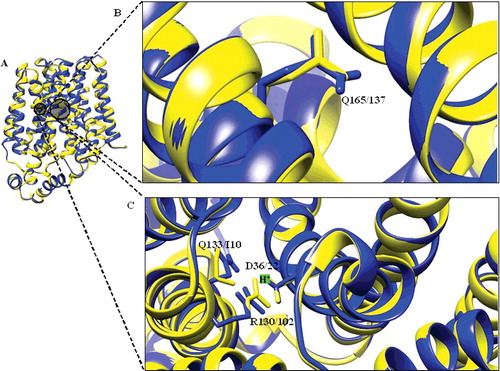
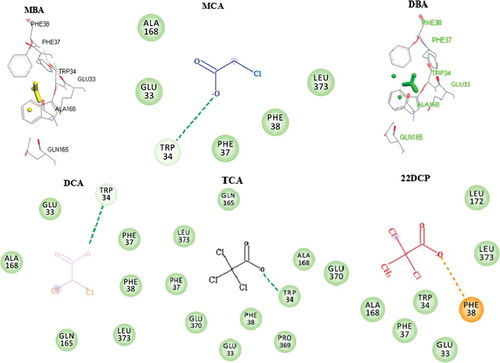
Figure 4. Docked conformation of DehrP (model) with DGlc. (A) Putative DehrP binding site in the inward-facing conformation (rainbow) complexed with D-glucose (DGlc), monobromoacetate (MBA), monochloroacetate (MCA), dibromoacetate (DBA), dichloroacetate (DCA), trichloroacetate (TCA) and 2,2-dichloropropionate (2,2-DCP) shown as rectangular grey shading. (B) Expanded view of the binding site residues Glu33, Trp34, Phe37, Phe38, Gln165 and Glu370 (stick model) and DGlc (purple), MBA (yellow), MCA (blue), DBA (green), DCA (pink), TCA (black) and 2, 2-DCP (red) in the ball and stick representation.
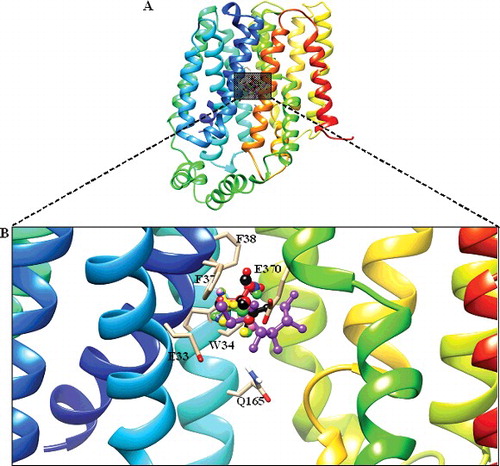
Figure 5. Docked conformation of GlcPse (template). (A) The glucose binding site in the inward-facing conformation (rainbow) complexed with D-glucose (DGlc) and monobromoacetate (MBA), monochloroacetate (MCA) shown as rectangular grey shading. (B) Expanded view of the binding site residues (Gln137, Gln250, Gln251, Asn256 and Trp367) complexed with DGlc (purple), MBA (yellow) and MCA (blue) in the ball and stick representation.
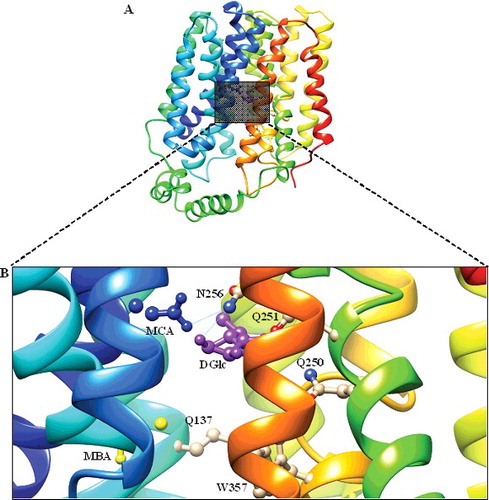
Figure 6. Illustration of halogen bonds and aromatic interactions in DehrP-haloacid complexes. The ligands and binding site residues are shown in the ball and stick presentation. The protein is presented as a cartoon model and coloured by atom type with carbon atom in grey, hydrogen in white, oxygen in red, nitrogen in blue, chloride in green and bromide in purple. (A) Monobromoacetate (MBA in yellow) ligand forming Br···aromatic ring interaction with Trp34. (B) Monochloroacetate (MCA in blue) ligand forming Cl···aromatic ring interaction with Phe38. (C) Dibromoacetate (DBA in green) ligand forming Br···aromatic ring interactions with Trp34 and Phe38. (D) Dichloroacetate (DCA in pink) ligand forming Cl···aromatic ring interactions with Trp34 and Phe38. (E) Trichloroacetate (TCA in black) ligand forming Cl···aromatic ring interactions with Trp34, Phe37 and Phe38. (F) 2,2-dichloropropionate (2,2-DCP in red) ligand forming Cl···aromatic ring interactions with Trp34 and Phe38.
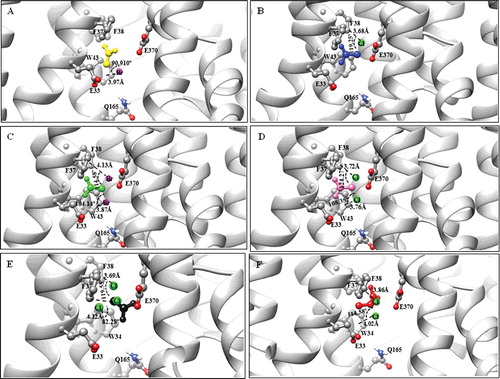
Figure 8. Proposed mechanism of haloacid:H+ Symport. (A) Transport cycle of DehrP shows the postulated steps with the outward-facing (I), and inward-facing (II) conformations separated by the occluded intermediate conformation (III). Haloacid (S; 2,2-DCP) and proton (H+; +) are coupled in the process. (B) Salt bridge forms between the side chains of Asp36 (-COO−) and Arg130 (-NH3+) in the absence of H+ (IV). Protonated D36 shifts closer to the Asp36 binding cavity (V). H+ and S are coordinated by the binding residues in the cavity (VI). The genomic localization of dehrP with dehalogenase genes (dehD, dehL and dehE) and their upstream promoters (P1 and P2) was drawn from [Citation15,Citation61].
![Figure 8. Proposed mechanism of haloacid:H+ Symport. (A) Transport cycle of DehrP shows the postulated steps with the outward-facing (I), and inward-facing (II) conformations separated by the occluded intermediate conformation (III). Haloacid (S; 2,2-DCP) and proton (H+; +) are coupled in the process. (B) Salt bridge forms between the side chains of Asp36 (-COO−) and Arg130 (-NH3+) in the absence of H+ (IV). Protonated D36 shifts closer to the Asp36 binding cavity (V). H+ and S are coordinated by the binding residues in the cavity (VI). The genomic localization of dehrP with dehalogenase genes (dehD, dehL and dehE) and their upstream promoters (P1 and P2) was drawn from [Citation15,Citation61].](/cms/asset/f9afb628-cd71-45ba-b0ef-235a7364d633/tbeq_a_1432417_f0008_oc.jpg)