Figures & data
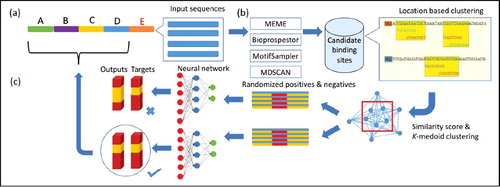
Figure 2. Stacked autoencoder neural network architecture. It consists of 76 input neurons in the input layer, 25 and 15 neurons in the first and second hidden layer, respectively. The output layer has two output neurons which represent motif and non-motif class; b is bias neuron.
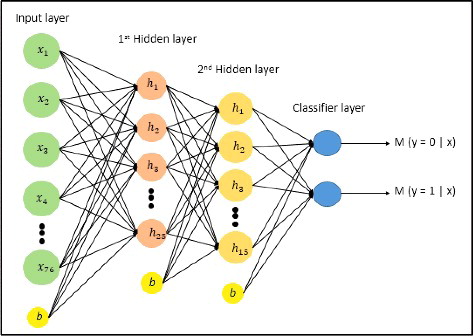
Table 1. Statistics of ten datasets used in this study.
Figure 3. Visual description of true positives (TP), false positives (FP), false negatives (FN) and true negatives (TN) on binding and non-binding regions in prediction. The black region indicates binding sites and the blue stretch indicates DNA region.
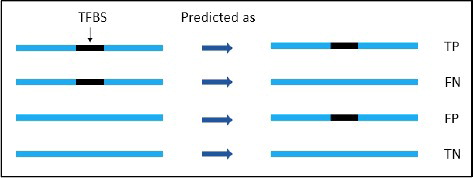
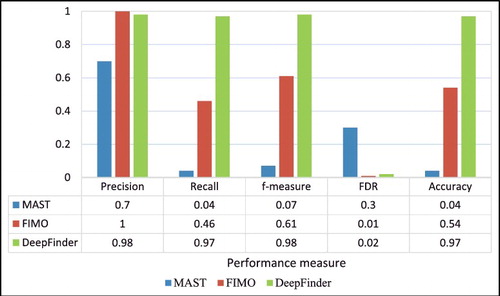
Table 2. Comparison of average precision and the recall and f-measure rates of MAST, FIMO and DeepFinder (DF) using five-fold cross validation.
Table 3. Comparison of false discovery rate (FDR), accuracy, and Matthews correlation coefficient (MCC) of MAST, FIMO, and DeepFinder (DF).
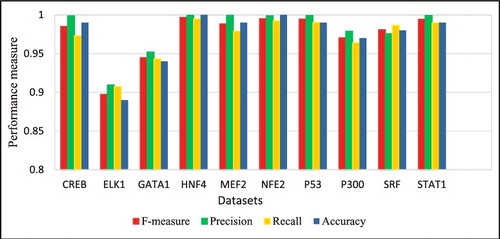
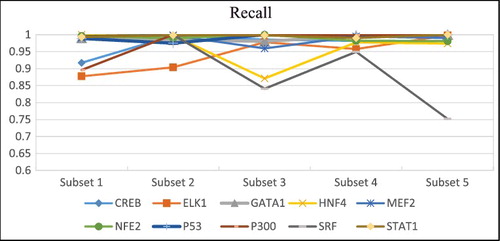
Figure 4. Average f-measure, precision, recall and accuracy rates obtained by DeepFinder on the ten datasets using five-fold cross-validation.