Figures & data
Figure 1. Photographs of eight varieties of Catharanthus roseus. (a) Patricia White (PW); (b) First Kiss Polka Dot (FKD); (c) First Kiss Peach (FKP); (d) Experimental Rose Pink (ER); (e) Experimental Deep Pink (ED); (f) Cooler Orchid (CO); (g) Victory Red (VR); (h) Blue Pearl (BP). Note: The colourless and coloured flowers are red and blue framed, respectively.
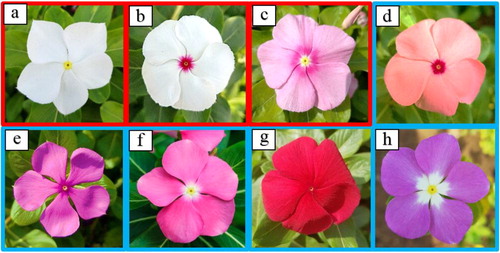
Table 1. Characteristics of eight varieties of Catharanthus roseus.
Table 2. Summary of alignment results to the chloroplast reference genome (GenBank No. KC561139).
Figure 2. Number of total and unique SNPs detected for each C. roseus variety. Note: More information about variety abbreviations is provided in .
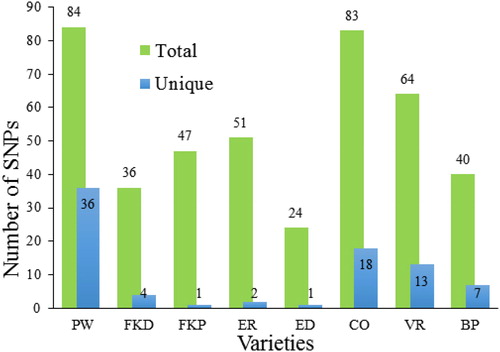
Figure 3. Locations of SNPs in the chloroplast genome. IGS, intergenic spacer; NS, non-synonymous; S, synonymous; I, intron; tRNA, tRNA-related sequence; rRNA, rRNA-related sequence. Note: More information about variety abbreviations is provided in .
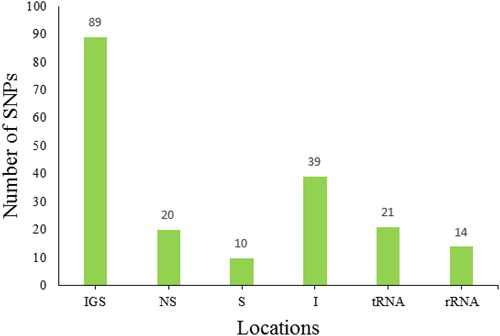
Table 3. Unique SNPs and their positions in the chloroplast genome.
Table 4. Summary of SNPs hotspots analysis results.
Table 5. The total of chloroplast DIPs ordered by their position in the genome.
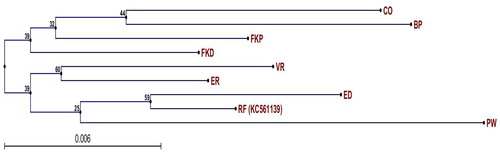
Figure 4. Neighbour-joining trees with bootstrap for chloroplast SNPs in eight C. roseus varieties. CO, Cooler Orchid; BP, Blue Pearl; FKP, First Kiss Peach; FKD, First Kiss Polka Dot; VR, Victory Red; ER, Experimental Rose Pink; ED, Experimental Deep Pink; RF, Reference plastome (GenBank No. KC561139); PW, Patricia White. Note: Numbers near each node are bootstraps used to derive estimates of standard errors and confidence intervals and the numbers towards the end of each branch represent dNS/dS values (dNS = non-synonymous substitution; dS = synonymous substitution).