Figures & data
Table 1. Overview of small RNA sequences for two wheat (Triticum aestivum L. cv. XF 20) libraries.
Table 2. Differentially expressed miRNA genes in wheat (Triticum aestivum L. cv. XF 20) in response to drought stress.
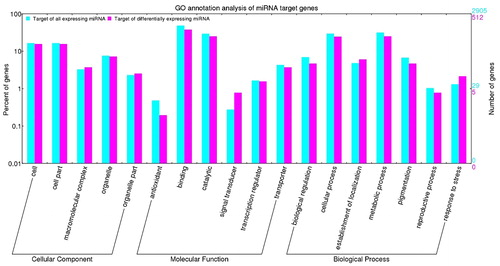
Figure 1. Gene ontology analysis of miRNA target genes identified in drought stress of wheat (Triticum aestivum L. cv. XF 20). Note: The digits on the left y-axis and right y-axis show the percentage and number of miRNA targets, respectively. Targets of all expressed miRNAs are shown in cyan; targets of all differentially expressed miRNAs are shown in pink.
Data availability
The Triticum aestivum (cv. XF 20) small RNA sequences, degradome sequences, and transcriptome sequences have all been deposited in the NCBI SRA database with accession number SRP 166912.
