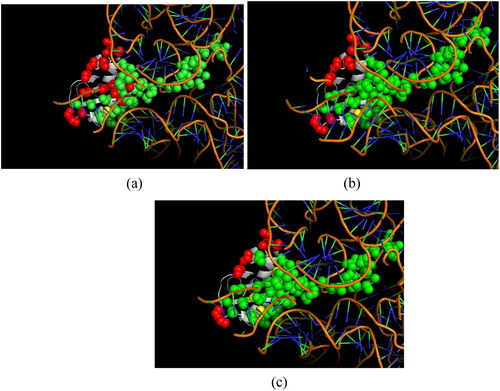
Figures & data
Table 1. The one-hot binary encoding for 20 types of amino acids.
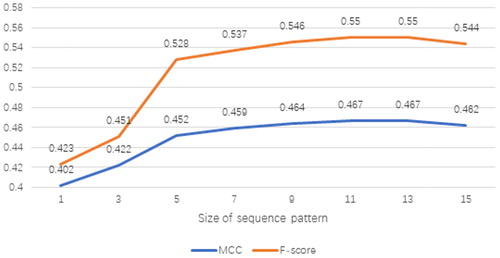
Figure 4. MCC and F-score with respect to different sizes of adjustment windows on validation set RB86.
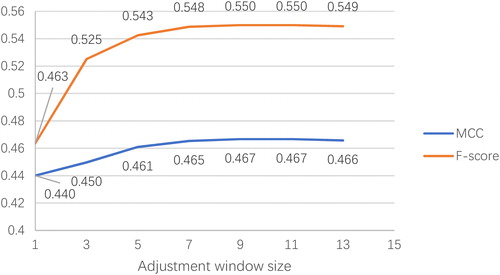
Table 2. Prediction performance of single and ensemble predictors on validation set RB86.
Table 3. Prediction performance comparison on testing set RB44.