Figures & data
Table 1. Primers used for cloning.
Figure 1. Construction of recombinant expression vector pET28a-Lac-enhanced green fluorescence protein (EGFP).
Note: EGFP fragment was cloned from pEGFP-N1 and ligated into pET28a vector to construct the intermediate vector pET28a-EGFP. pUC18 vector was used as the template to introduce Lac promoter to obtain recombinant vector pET28a-Lac-EGFP.
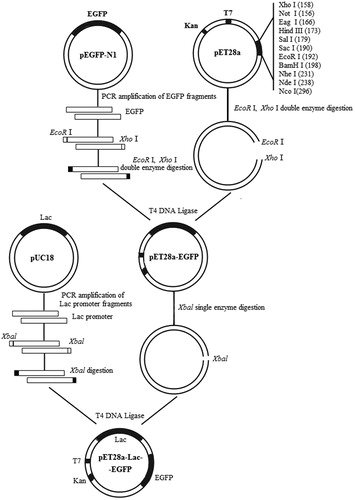
Figure 2. Map of the constructed recombinant vector pET28a-Lac-enhanced green fluorescence protein (EGFP).
Note: It is a closed 6138-bp long loop structure and includes a T7 promoter, an inserted Lac fragment, a kanamycin-resistant fragment, an EGFP-encoding region and enzyme cleavage sites such as EcoRI, XhoI and XbaI.
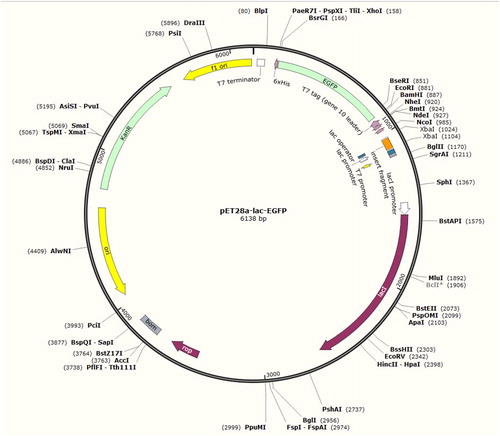
Figure 3. Electrophoresis plot of vector pET28a-Lac-EGFP (a) and a nucleotide sequence map of pET28a-Lac-enhanced green fluorescence protein (EGFP) vector (b).
Note: (a) Lane 1-3, products digested by restriction enzymes for recombinant plasmid pET28a-Lac-EGFP; Lane M, DNA marker. (b) Underlined portion is fragment of Lac promoter with 74 bp size; the fragment highlighted in yellow is ribosome-binding site (RBS); EGFP-encoding sequence is highlighted in purple; starter code is ATG.
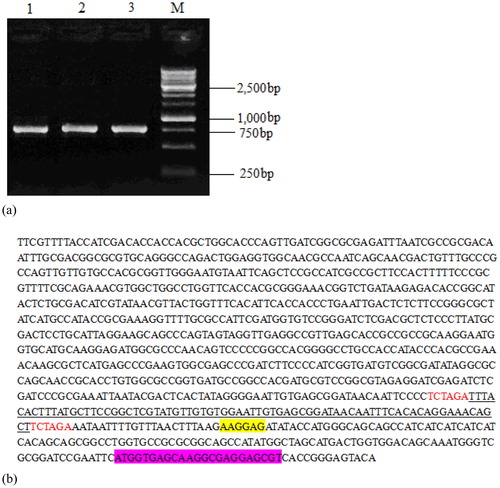
Figure 4. Morphological observation of positive transformants under natural light (a) and under UV light in a dark room (b,c) without IPTG induction (b) and under IPTG induction (c).
Note: (a) Colonial morphology of positive transformants under natural light conforming to basic morphological characteristics of Klebsiella variicola. (b) No obvious green fluorescence without IPTG induction under the UV lamp in the dark. (c) Green fluorescence in positive transformants under IPTG induction, observed under the UV lamp in the dark.
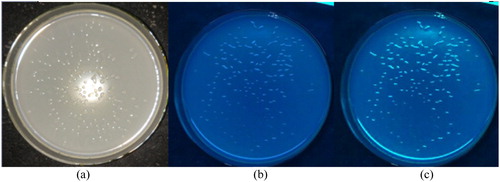
Figure 5. SDS-PAGE of EGFP slightly expressed in Klebsiella variicola GN02.
Note: Black arrow is target band OF approximately 26.9 kDa. Lanes 1 and 3, positive transformants without isopropyl β-D-1-thiogalactopyranoside (IPTG) induction; Lanes 2 and 4, IPTG-induced positive transformants.
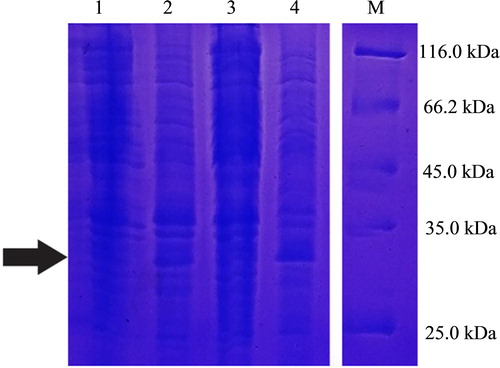
Figure 6. Promotion of Pennisetum sinense growth induced by Klebsiella variicola GN02 after 5 days. Note: (a) Red arrow indicates where bacteria congregated. (b) Inoculation with K. variicola GN02 strain effectively increased the number of roots, especially fibrous roots, and their root surface area, p < 0. 05.
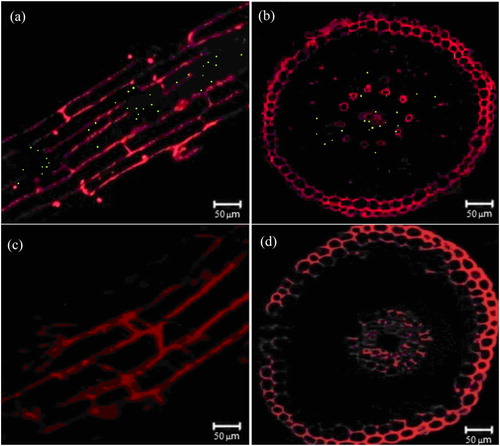
Figure 7. Observation of Pennisetum sinense with and without inoculation of EGFP-labelled Klebsiella variicola GN02 under laser confocal scanning microscope. Longitudinal (a) and cross cuts (b) of green fluorescent dots in P. sinense roots inoculated with EGFP-labelled K. variicola GN02. Longitudinal (c) and cross cuts (d) of P. sinense roots not inoculated with EGFP-labelled K. variicola GN02.