Figures & data
Table 1. List of primers used in DD-PCR and total number of up/down-regulated bands.
Figure 1. Agarose gel electrophoresis of RT-PCR detection (A) of Tobacco mosaic virus-coat protein (TMV-CP) and DD-PCR (B to F) using primers A2, A4, RAPD 9, At1 and At5. Lane Marker: DNA marker 100 bp ladder, lane “Mock” control (mock-inoculated plant tissues), lanes 6 dpi, 9 dpi and 15 dpi: plant tissues collected at 6, 9 and 15 dpi of TMV. White round rectangle indicates the bands that were selected for sequencing. Note: Agarose gel electrophoresis (1.5%) in TBE buffer.
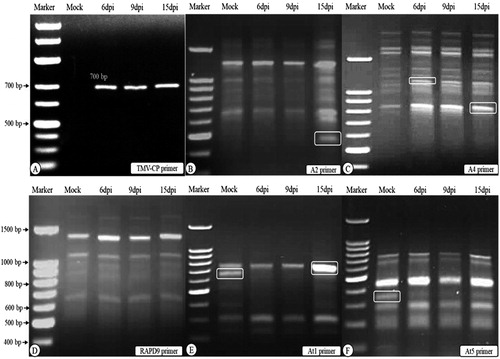
Figure 2. Phylogenetic tree (A) constructed based on the obtained DNA sequences of the chloroplast isolated genes in this study and the sequence alignment (B) between the two closely related chloroplast-associated genes (MG565980 and MG565978).

Figure 3. Diagram showing the predicted transmembrane domains of the deduced amino-acid sequences of the isolated genes.
Figure 4. Phylogenetic analysis showing the genetic relationship between the nucleotide sequence of the tomato (S. lycopersicum) kinesin-like protein gene (MG565981) and other kinesin-like protein genes available in GenBank. The phylogeny was tested with 2000 bootstrap replicates based on the UPGMA statistical method.
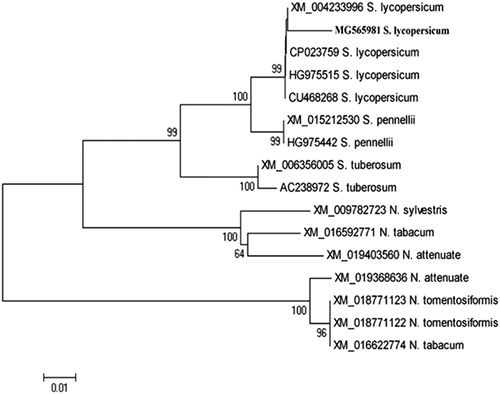
Figure 5. DNA nucleotide sequence of tomato kinesin-like protein gene. The deduced amino acid sequence of the protein is shown below the DNA sequence. Potential Protein kinase C phosphorylation site (three aa), Casein kinase II Phosphorylation site (four aa) and N-myristoylation site (six aa) are shadowed and underlined. Black colour indicates interaction between two sites.
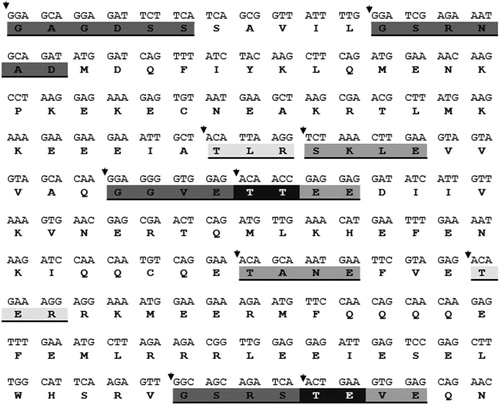
Figure 6. Phylogenetic analysis showing the genetic relationship between nucleotide sequence of the kinesin-like protein gene (MG565981) and other 18 defensin genes available in GenBank. The kinesin had high similarity to the four defencing genes isolated from Arabidopsis plant. The phylogeny tested with bootstrap method with 2,000 replications and generated based on UPGMA statistical method.