Figures & data
Table 1. Simple sequence repeat (SSR) types in the carrot genome.
Table 2. Most abundant simple sequence repeat (SSR) motifs.
Table 3. Statistics on simple sequence repeat (SSR) marker numbers and locations in the carrot genome assembly.
Table 4. Statistics of cross-species marker transferability to fennel genome assembly.
Figure 1. A set of polymorphic marker fragments visualized by high-resolution capillary electrophoresis. Electropherograms of different size alleles amplified from carrot landraces are displayed for each marker. Primer sequences of markers can be accessed at https://figshare.com/articles/Marker_data/8593337/2.
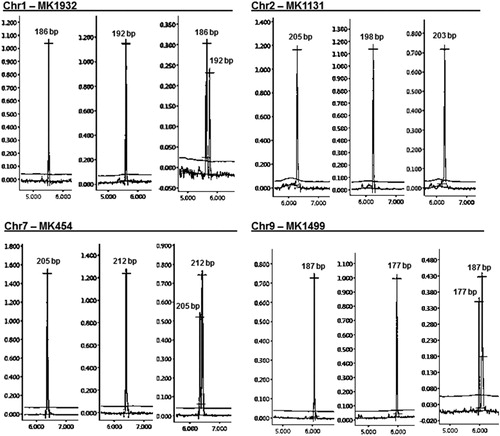
Table 5. Informativeness of the codominant, polymorphic markers.
Data availability statement
All data generated during this study are available at Figshare (https://doi.org/10.6084/m9.figshare.8593337).
