Figures & data
Figure 1. Comparison of common structural indexes in PARP-1 in four different species. Note: H = Human, B = Bovine, M = Mouse, R = Rat.
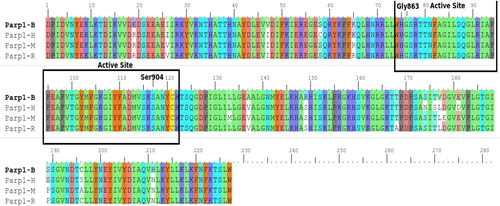
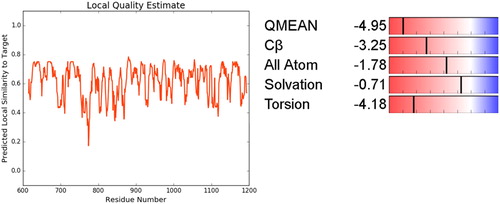
Figure 6. Ramachandran plot for PARP4, Ramachandran contour plot depicting Φ and Ψ angles. Source: http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php.
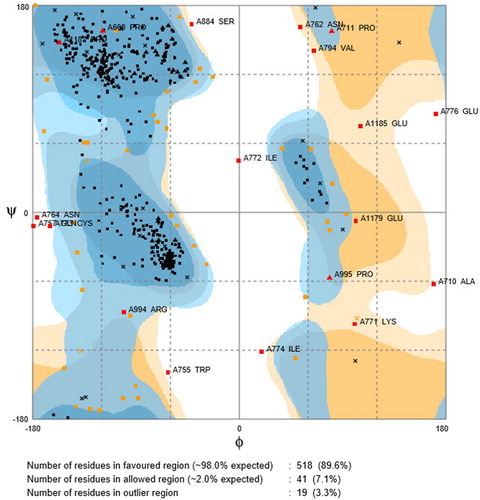
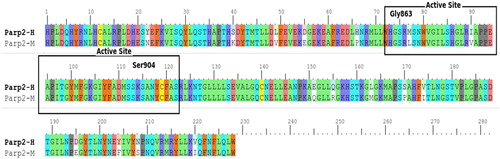
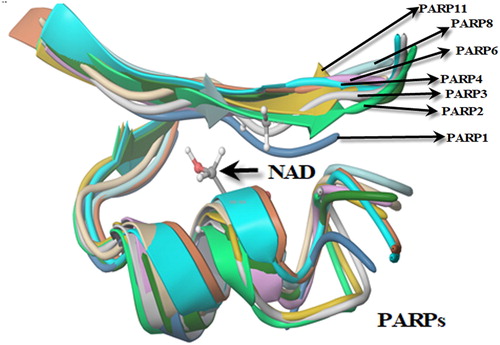
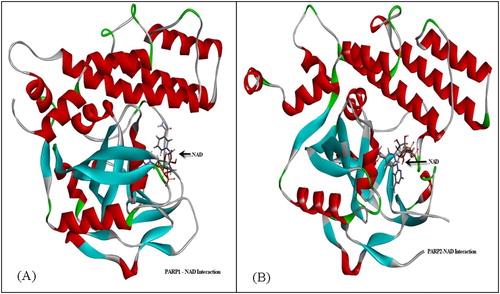
Figure 10. Active site determination based on common structural motif that recognizes the NAD region of PARPs belonging to human, rat and mouse.
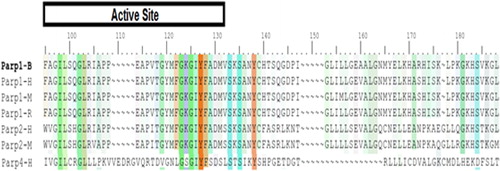
Table 1. Comparative RMSD values of PARP1, PARP and PARP4 (°A).
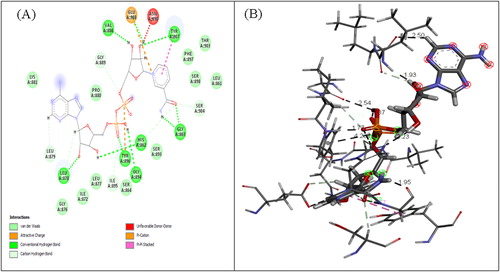
Table 2. Catalytic sites from PARPs with the highest probability of interaction with NAD.
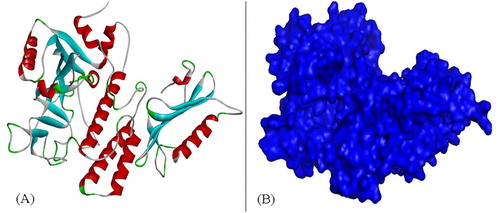
Figure 13. Electrophoresis (A) and chromatography (B) analysis of PARP1 and PARP2. Note: For electrophoresis analysis, 5 μg protein was loaded to gel.
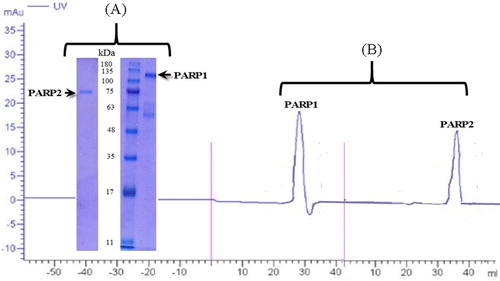
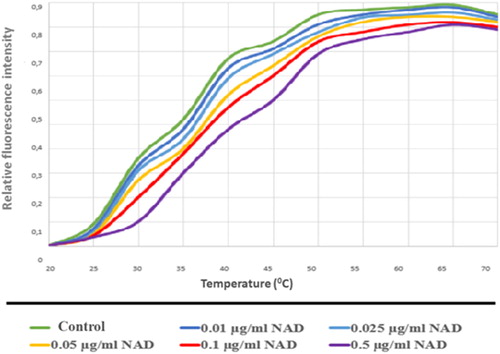
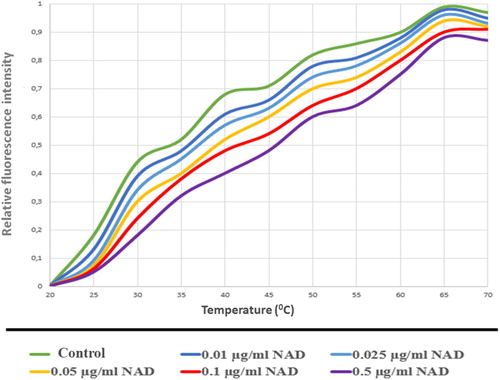
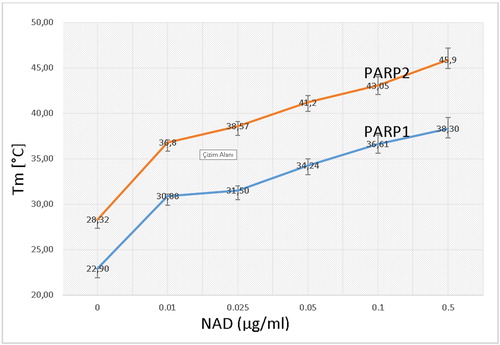
Figure 14. PARP1-NAD interaction was tested with thermal shift assay. Melting temperatures of PARP1-NAD interaction (Tm values).