Figures & data
Figure 1. Enrichment of anti-IL1RAP scFvs by phage panning technique. (A) Silver staining for biotinylated IL1RAP protein. Note: Purchased human IL1RAP proteins were biotinylated and enriched by magnetic beads coated with streptavidin, and the supernatant was collected. After a wash, the biotinylated protein was eluted from beads. Each sample was checked by SDS-PAGE electrophoresis followed by silver staining. (B) Enrichment analysis of phage library screening for IL1RAP scFv. (C) Specific scFv enrichment analysis by ELISA. Note: IL1RAP proteins were coated and the phages population was added into the wells to detect their binding. The absorbance was read at OD405.
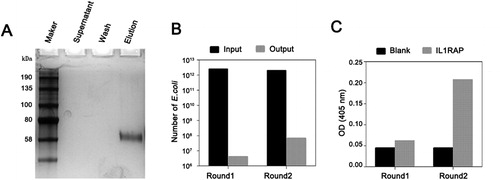
Figure 2. Selection of yeast displayed anti-IL1RAP scFv by flow sorting. (A) A yeast-display IL1RAP scFv library enriched for scFv binding to 100 nmol/L of biotinylated IL1RAP protein by first round of flow sorting. (B) The second round of selection for scFv binding to 50 nmol/L of biotinylated IL1RAP protein. (C) The third round of selection for scFv binding to 10 nmol/L of biotinylated IL1RAP protein. Note: IL1RAP binding to yeast-display scFv was measured by staining with biotinylated IL1RAP protein and anti-c-Myc antibody detected by PE-labeled streptavidin (or APC-labeled neutravidin) and FITC-labeled secondary antibody (c, f, i). As negative controls, yeast cells were not stained (a, d, g) and incubate without antigen (b, h) or biotinylated Her2 protein (e).
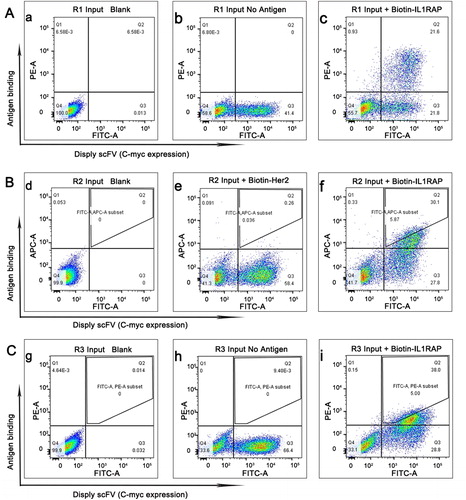
Figure 3. Sequence analysis and binding affinity test of selected positive scFvs. (A) Sequence alignment of scFv-02 and scFv-61. Alignments indicating highly conserved sequences are shown in colors. The CDRs of the variable domains are indicated. (B) Structural configuration of scFv-02 and scFv-61 by protein modeling software Swiss-model. (C) Analysis of anti-IL1RAP scFv binding by flow cytometry. Note: IL1RAP binding to yeast-display scFv was measured by staining with 5 nmol/L of biotinylated IL1RAP protein. FITC staining to detect the Myc tag (scFv-Myc) and SA-PE staining to detect the biotinylated IL1RAP antigen.
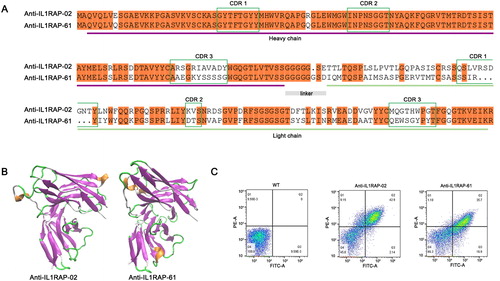
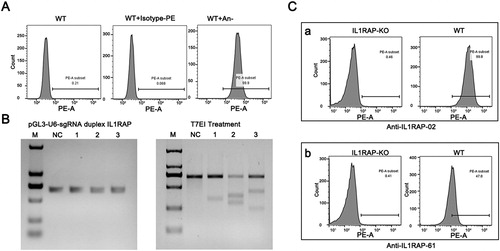
Figure 4. Binding specificity of selected scFvs with IL1RAP antigen on 293F cell surface. (A) FACS analysis of endogenous expression of IL1RAP on 293F cell surface. Purchased PE-labeled anti-IL1RAP antibodies were used to detect the endogenous expression of IL1RAP on 293F cell surface. (B) T7EI assay to confirm the efficiency of CRISPR-Cas9 mediated IL1RAP editing. (C) Cell binding assay to verify the specificity of scFv-02 and scFv-61 to IL1RAP antigen on 293F cell surface.