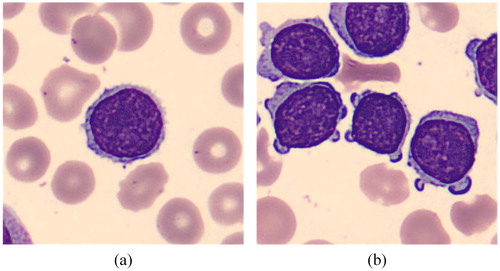
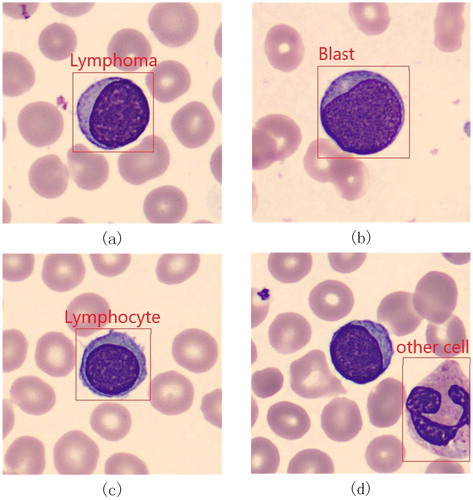
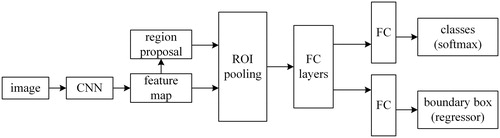
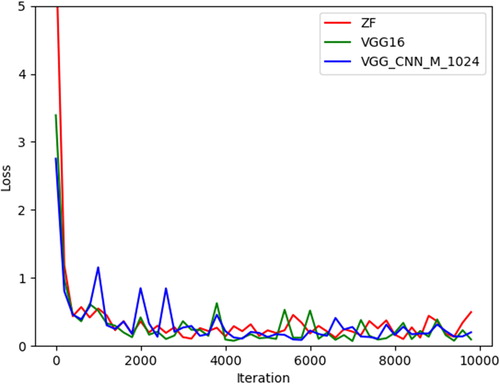
Figures & data
Table 1. Total number of cells of each class in the dataset.
Table 2. Number of images of each part.
Table 3. mAP of 6 combinations of different methods and networks.
Table 4. Confusion matrix of the final model.
Table 5. Confusion matrix of KNN.
Table 6. Confusion matrix of VGG16.
Data availability statement
Raw data were generated at East China Normal University. Derived data supporting the findings of this study are available from the corresponding author Qingli Li on request. More details can be found from http://bio-hsi.ecnu.edu.cn