Figures & data
Figure 1. Typical nut characteristics for Juglans regia (a), J. sigillata (b), J. sigillata × J. regia (c), and J. hopeiensis (d).

Table 1. Walnut cultivars (Juglans regia and J. sigillata) used for the AFLP analysis.
Table 2. Summary of the AFLP markers scored for J. regia, J. sigillata and the hybrids using 9 primer combinations.
Table 3. Na, Ne, H, and I for J. regia, J. sigillata and the hybrids based on AFLP analysis.
Figure 2. Factorial correspondence analysis (FCA) based on polymorphisms at AFLP loci for 35 walnut cultivars.
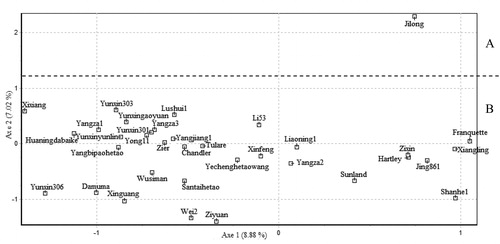
Figure 3. Dendrogram of 35 walnut cultivars based on AFLP analysis conducted using 9 primer combinations.
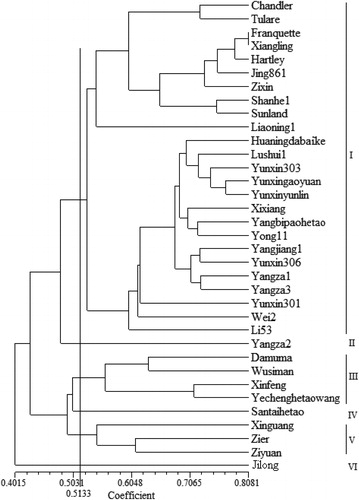
Table 4. Nei’s genetic identity (above diagonal) and genetic distance (below diagonal) among 4 groups.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
