Figures & data
Figure 1. Multiple alignment of the deduced amino acid sequences of the putative protein encoded by the BrERF11b gene with those of selected ERF transcription factors. The unrooted phylogenetic tree of ERF transcription factor was constructed using neighbor-joining method with 1000 bootstrap replicates. Bootstrap values are indicated at the branches. The GenBank accession number is shown following the abbreviation of the ERF transcription factor in the unrooted phylogenetic tree.
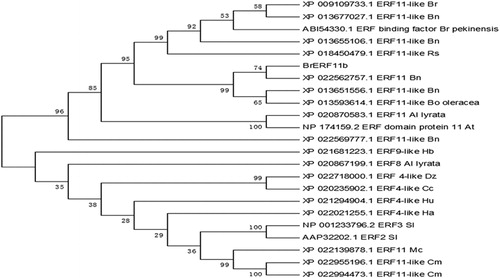
Figure 2. Southern blot analysis of the transgenic plants. (A) lane 1, WT Nicotiana bethamiana; lane 2, plasmid with BrERF11b gene; lane 3, WT N, tabacum variety K326; lane 4, BLine25; lane 5, KLine13; lane 6, BLine26; lane 7, Kline12 (DNA in lanes 1 to 7 was digested by EcoR I); lane 8, WT Nicotiana bethamiana; lane 9, BLine25; lane 10, BLine26; lane 11, the plasmid with BrERF11b gene, (DNA in lanes 8 to 11 was digested by Xba I). (B) lane 1, the plasmid with BrERF11b gene; lane 2, WT N. tabacum variety K326;lane 3, KLine12; lane 4, KLine13; lane 5, KLine15; lane 6, KLine18 (DNA in lanes 1 to 6 was digested by Pst I). Note: Genomic DNA extracted from the transgenic plant using CTAB reagent was digested by restriction enzyme, and separated in an 0.8% agarose gel, and then transferred onto Hybond-N + membrane (Amerasham). Individual lanes were hybridized with dioxigenin (DIG)-11-dUTP–labeled cDNA probes specific for the segment of the BrERF11b gene (the putative open reading frame regions).
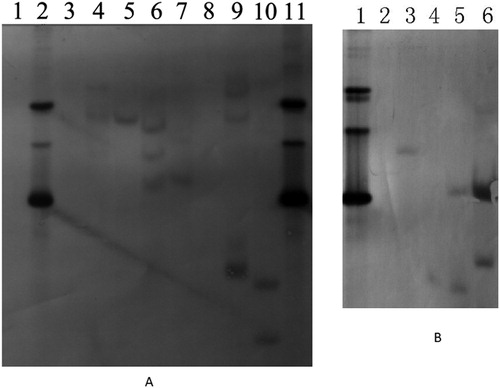
Figure 3. RT-PCR of transgenic plants. (A) RT-PCR of plants from transgenic plant lines KLine13 and KLine15. lane1, DNA Marker (DL10000 DNA Marker, Takara Bio Inc., Beijing.); lanes 2, 3 and 4, KLine13; lanes 5, 6 and 7, KLine15; lane 8, WT N. tabacum variety K326. (B) RT-PCR of plants from transgenic plant lines BLine25 and BLine26. lanes 1, 2 and 3, BLine25; lane 4, DNA Marker (DL10000 DNA Marker, Takara Bio Inc., Beijing); lane 5, WT N. benthamiana; lanes 6,7 and 8, BLine26. (C) RT-PCR of plants from transgenic plant line KLine12. lanes 1, 2, 3, 4 and 6, KLine12; lane 5, DNA Marker (GeneRuler 1 Kb DNA Ladder, Thermo Fisher Scientific Company, China); lane 7, plasmid with BrERF11b gene; lane 8, WT N. tabacum variety K326.
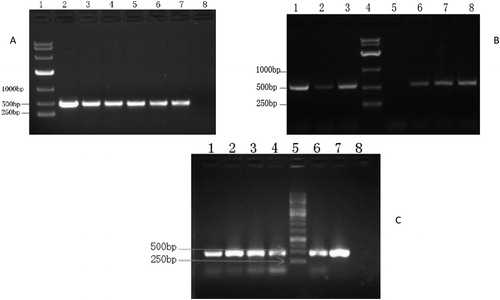
Figure 4. Resistance of transgenic plants to the larvae of Spodoptera litura. (A) Weight of S. litura larvae feeding on the leaves of the transgenic plant line KLine12; (B) Survival rate of S. litura feeding on the leaves of the transgenic plant line KLine12; (C): Weight of S. litura larvae feeding on the leaves of the transgenic plant lines KLine13 and KLine15; (D) Weight of S. litura larvae feeding on the leaves of the transgenic plant lines BLine 25 and BLine 26. Note: Data are mean values with standard deviation (±SD) from eight replicate experiments. Duncan's or Student’s test was used to test for significance (p < 0.05), indicated by different lowercase letters.
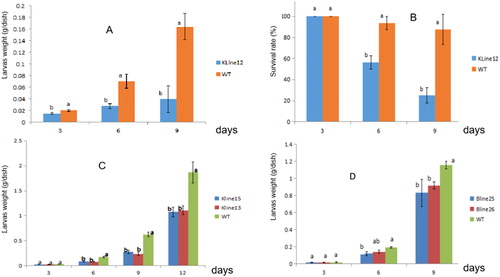
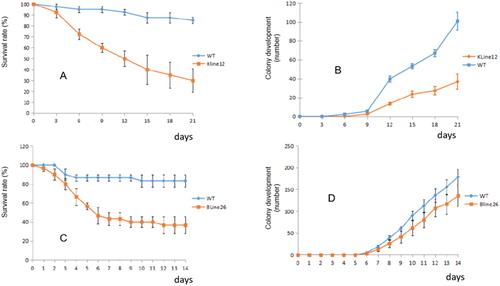
Figure 5. Resistance of transgenic plants to the green aphids. (A) Survival rate of the green aphid feeding on the leaves of the transgenic plant line KLine12; (B) Colony development of the green aphid feeding on the leaves of the transgenic plant line KLine12; (C) Survival rate of the green aphid feeding on the leaves of the transgenic plant line BLine26; (D) Colony development of the green aphid feeding on the leaves of the transgenic plant line BLine26.
Supplemental Material
Download PDF (645.2 KB)Data availability statement
The data supporting this paper are available from the corresponding author upon reasonable request.
