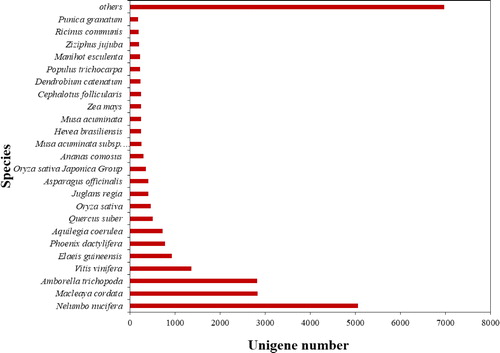
Figures & data
Table 1. Schisandra chinensis accessions analyzed in this work.
Table 2. Functional annotation of S. chinensis unigenes.
Table 3. Type, number and frequency of expressed sequence tag-simple sequence repeat markers in Schisandra chinensis.
Table 4. Distribution of EST-SSR motifs in the transcriptome of S. chinensis.
Figure 2. Frequency distribution of putative EST-SSRs identified from S. chinensis ESTs, according to motif type (a) and number of repeats (b).
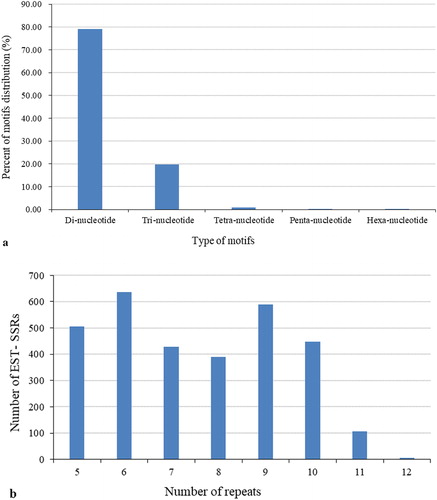
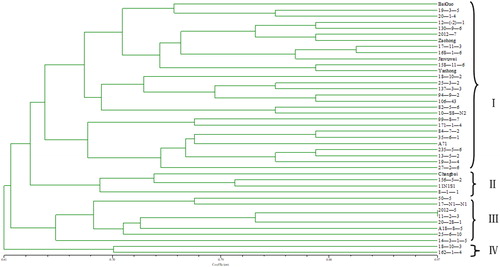
Table 5. Information of 14 SSRs used for assessment of genetic diversity among 42 S. chinensis accessions.
Table 6. Estimation of genetic diversity based on 14 SSRs among 42 S. chinensis.
Availability of data and materials
The raw data of RNA-seq are deposited in Sequence Read Archives Database (http://www.ncbi.nlm.nih.gov/bioproject/609148) under accession number PRJNA609148. Other datasets supporting the conclusions of this article are included within the article (and its additional file).